About the Execution of Marcie for MAPKbis-PT-5320
| Execution Summary | |||||
| Max Memory Used (MB) |
Time wait (ms) | CPU Usage (ms) | I/O Wait (ms) | Computed Result | Execution Status |
| 5449.735 | 4734.00 | 4000.00 | 60.00 | FTTTTFFFTTTFTTFT | normal |
Execution Chart
We display below the execution chart for this examination (boot time has been removed).
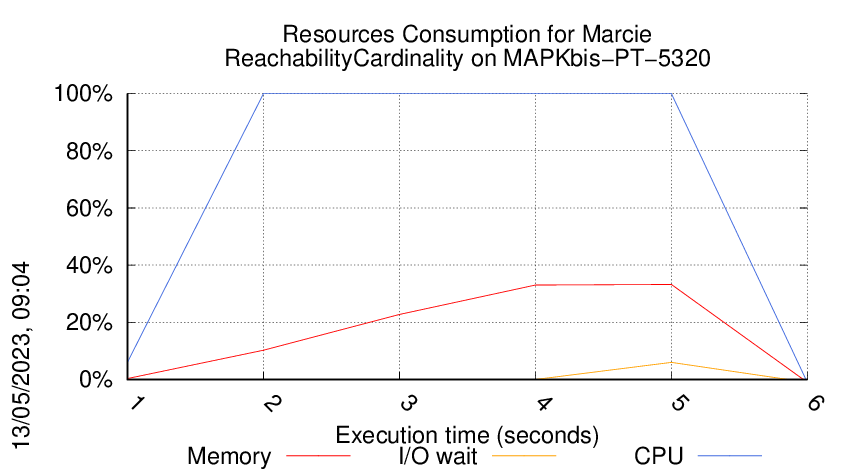
Trace from the execution
Formatting '/data/fkordon/mcc2023-input.r225-tall-167856407900646.qcow2', fmt=qcow2 size=4294967296 backing_file=/data/fkordon/mcc2023-input.qcow2 cluster_size=65536 lazy_refcounts=off refcount_bits=16
Waiting for the VM to be ready (probing ssh)
..........................
=====================================================================
Generated by BenchKit 2-5348
Executing tool marcie
Input is MAPKbis-PT-5320, examination is ReachabilityCardinality
Time confinement is 3600 seconds
Memory confinement is 16384 MBytes
Number of cores is 1
Run identifier is r225-tall-167856407900646
=====================================================================
--------------------
preparation of the directory to be used:
/home/mcc/execution
total 768K
-rw-r--r-- 1 mcc users 8.3K Feb 25 16:19 CTLCardinality.txt
-rw-r--r-- 1 mcc users 76K Feb 25 16:19 CTLCardinality.xml
-rw-r--r-- 1 mcc users 11K Feb 25 16:03 CTLFireability.txt
-rw-r--r-- 1 mcc users 47K Feb 25 16:03 CTLFireability.xml
-rw-r--r-- 1 mcc users 4.2K Jan 29 11:40 GenericPropertiesDefinition.xml
-rw-r--r-- 1 mcc users 6.4K Jan 29 11:40 GenericPropertiesVerdict.xml
-rw-r--r-- 1 mcc users 4.3K Feb 25 16:22 LTLCardinality.txt
-rw-r--r-- 1 mcc users 25K Feb 25 16:22 LTLCardinality.xml
-rw-r--r-- 1 mcc users 5.4K Feb 25 16:22 LTLFireability.txt
-rw-r--r-- 1 mcc users 21K Feb 25 16:22 LTLFireability.xml
-rw-r--r-- 1 mcc users 16K Feb 25 16:39 ReachabilityCardinality.txt
-rw-r--r-- 1 mcc users 131K Feb 25 16:39 ReachabilityCardinality.xml
-rw-r--r-- 1 mcc users 40K Feb 25 16:29 ReachabilityFireability.txt
-rw-r--r-- 1 mcc users 153K Feb 25 16:29 ReachabilityFireability.xml
-rw-r--r-- 1 mcc users 1.7K Feb 25 16:22 UpperBounds.txt
-rw-r--r-- 1 mcc users 3.8K Feb 25 16:22 UpperBounds.xml
-rw-r--r-- 1 mcc users 6 Mar 5 18:22 equiv_col
-rw-r--r-- 1 mcc users 5 Mar 5 18:22 instance
-rw-r--r-- 1 mcc users 6 Mar 5 18:22 iscolored
-rw-r--r-- 1 mcc users 170K Mar 5 18:22 model.pnml
--------------------
content from stdout:
=== Data for post analysis generated by BenchKit (invocation template)
The expected result is a vector of booleans
BOOL_VECTOR
here is the order used to build the result vector(from text file)
FORMULA_NAME MAPKbis-PT-5320-ReachabilityCardinality-00
FORMULA_NAME MAPKbis-PT-5320-ReachabilityCardinality-01
FORMULA_NAME MAPKbis-PT-5320-ReachabilityCardinality-02
FORMULA_NAME MAPKbis-PT-5320-ReachabilityCardinality-03
FORMULA_NAME MAPKbis-PT-5320-ReachabilityCardinality-04
FORMULA_NAME MAPKbis-PT-5320-ReachabilityCardinality-05
FORMULA_NAME MAPKbis-PT-5320-ReachabilityCardinality-06
FORMULA_NAME MAPKbis-PT-5320-ReachabilityCardinality-07
FORMULA_NAME MAPKbis-PT-5320-ReachabilityCardinality-08
FORMULA_NAME MAPKbis-PT-5320-ReachabilityCardinality-09
FORMULA_NAME MAPKbis-PT-5320-ReachabilityCardinality-10
FORMULA_NAME MAPKbis-PT-5320-ReachabilityCardinality-11
FORMULA_NAME MAPKbis-PT-5320-ReachabilityCardinality-12
FORMULA_NAME MAPKbis-PT-5320-ReachabilityCardinality-13
FORMULA_NAME MAPKbis-PT-5320-ReachabilityCardinality-14
FORMULA_NAME MAPKbis-PT-5320-ReachabilityCardinality-15
=== Now, execution of the tool begins
BK_START 1678641244686
bash -c /home/mcc/BenchKit/BenchKit_head.sh 2> STDERR ; echo ; echo -n "BK_STOP " ; date -u +%s%3N
Invoking MCC driver with
BK_TOOL=marcie
BK_EXAMINATION=ReachabilityCardinality
BK_BIN_PATH=/home/mcc/BenchKit/bin/
BK_TIME_CONFINEMENT=3600
BK_INPUT=MAPKbis-PT-5320
Not applying reductions.
Model is PT
ReachabilityCardinality PT
timeout --kill-after=10s --signal=SIGINT 1m for testing only
Marcie built on Linux at 2019-11-18.
A model checker for Generalized Stochastic Petri nets
authors: Alex Tovchigrechko (IDD package and CTL model checking)
Martin Schwarick (Symbolic numerical analysis and CSL model checking)
Christian Rohr (Simulative and approximative numerical model checking)
marcie@informatik.tu-cottbus.de
called as: /home/mcc/BenchKit/bin//../marcie/bin/marcie --net-file=model.pnml --mcc-file=ReachabilityCardinality.xml --memory=6 --mcc-mode
parse successfull
net created successfully
Net: MAPKbis_PT_5320
(NrP: 106 NrTr: 173 NrArc: 986)
parse formulas
formulas created successfully
place and transition orderings generation:0m 0.002sec
net check time: 0m 0.000sec
init dd package: 0m 2.876sec
RS generation: 0m 0.037sec
-> reachability set: #nodes 235 (2.4e+02) #states 8,126,465 (6)
starting MCC model checker
--------------------------
checking: AG [p70_equals_1<=0]
normalized: ~ [E [true U ~ [p70_equals_1<=0]]]
abstracting: (p70_equals_1<=0)
states: 8,126,465 (6)
-> the formula is TRUE
FORMULA MAPKbis-PT-5320-ReachabilityCardinality-03 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.000sec
checking: EF [p70_equals_0<=0]
normalized: E [true U p70_equals_0<=0]
abstracting: (p70_equals_0<=0)
states: 0
-> the formula is FALSE
FORMULA MAPKbis-PT-5320-ReachabilityCardinality-06 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.000sec
checking: AG [1<=AKT_0]
normalized: ~ [E [true U ~ [1<=AKT_0]]]
abstracting: (1<=AKT_0)
states: 8,126,465 (6)
-> the formula is TRUE
FORMULA MAPKbis-PT-5320-ReachabilityCardinality-08 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.000sec
checking: AG [TAK1_equals_0<=1]
normalized: ~ [E [true U ~ [TAK1_equals_0<=1]]]
abstracting: (TAK1_equals_0<=1)
states: 8,126,465 (6)
-> the formula is TRUE
FORMULA MAPKbis-PT-5320-ReachabilityCardinality-09 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.000sec
checking: EF [~ [RAF_equals_1<=p38_equals_1]]
normalized: E [true U ~ [RAF_equals_1<=p38_equals_1]]
abstracting: (RAF_equals_1<=p38_equals_1)
states: 8,126,465 (6)
-> the formula is FALSE
FORMULA MAPKbis-PT-5320-ReachabilityCardinality-11 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.000sec
checking: EF [~ [[Apoptosis_equals_0<=1 | TGFBR_equals_1<=0]]]
normalized: E [true U ~ [[Apoptosis_equals_0<=1 | TGFBR_equals_1<=0]]]
abstracting: (TGFBR_equals_1<=0)
states: 8,126,465 (6)
abstracting: (Apoptosis_equals_0<=1)
states: 8,126,465 (6)
-> the formula is FALSE
FORMULA MAPKbis-PT-5320-ReachabilityCardinality-14 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.000sec
checking: AG [PDK1_equals_1<=FOS_equals_1]
normalized: ~ [E [true U ~ [PDK1_equals_1<=FOS_equals_1]]]
abstracting: (PDK1_equals_1<=FOS_equals_1)
states: 8,126,465 (6)
-> the formula is TRUE
FORMULA MAPKbis-PT-5320-ReachabilityCardinality-15 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.000sec
checking: AG [[[[[[~ [MDM2_equals_1<=1] | ~ [[[[PLCG_equals_1<=Apoptosis_equals_0 | PKC_equals_1<=0] & ~ [1<=p21_equals_1]] & [~ [p53_equals_0<=0] | [DNA_damage_equals_1<=0 & EGFR_equals_0<=1]]]]] & [1<=Growth_Arrest_equals_1 | ~ [[~ [PDK1_equals_1<=0] | [~ [TAOK_equals_1<=1] & ~ [EGFR_stimulus_equals_1<=RAS_equals_1]]]]]] | ~ [1<=MYC_equals_0]] | ~ [1<=MAX_equals_1]] | 1<=FOS_equals_0]]
normalized: ~ [E [true U ~ [[1<=FOS_equals_0 | [~ [1<=MAX_equals_1] | [~ [1<=MYC_equals_0] | [[1<=Growth_Arrest_equals_1 | ~ [[[~ [EGFR_stimulus_equals_1<=RAS_equals_1] & ~ [TAOK_equals_1<=1]] | ~ [PDK1_equals_1<=0]]]] & [~ [[[[DNA_damage_equals_1<=0 & EGFR_equals_0<=1] | ~ [p53_equals_0<=0]] & [~ [1<=p21_equals_1] & [PLCG_equals_1<=Apoptosis_equals_0 | PKC_equals_1<=0]]]] | ~ [MDM2_equals_1<=1]]]]]]]]]
abstracting: (MDM2_equals_1<=1)
states: 8,126,465 (6)
abstracting: (PKC_equals_1<=0)
states: 8,126,465 (6)
abstracting: (PLCG_equals_1<=Apoptosis_equals_0)
states: 8,126,465 (6)
abstracting: (1<=p21_equals_1)
states: 4,063,232 (6)
abstracting: (p53_equals_0<=0)
states: 4,194,304 (6)
abstracting: (EGFR_equals_0<=1)
states: 8,126,465 (6)
abstracting: (DNA_damage_equals_1<=0)
states: 0
abstracting: (PDK1_equals_1<=0)
states: 8,126,465 (6)
abstracting: (TAOK_equals_1<=1)
states: 8,126,465 (6)
abstracting: (EGFR_stimulus_equals_1<=RAS_equals_1)
states: 8,126,465 (6)
abstracting: (1<=Growth_Arrest_equals_1)
states: 4,063,232 (6)
abstracting: (1<=MYC_equals_0)
states: 4,063,233 (6)
abstracting: (1<=MAX_equals_1)
states: 4,063,232 (6)
abstracting: (1<=FOS_equals_0)
states: 8,126,465 (6)
-> the formula is TRUE
FORMULA MAPKbis-PT-5320-ReachabilityCardinality-02 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.003sec
checking: AG [[[[~ [MTK1_equals_1<=DUSP1_equals_1] & [[[1<=Apoptosis_equals_0 & ~ [1<=Growth_Arrest_equals_1]] & [MYC_equals_1<=Growth_Arrest_equals_1 | 1<=SPRY_equals_0]] | ~ [[[[[MAP3K1_3_equals_1<=FGFR3_stimulus_equals_0 | 1<=ATM_equals_1] | ~ [TGFBR_stimulus_equals_0<=1]] & ~ [[1<=MEK1_2_equals_1 & MAP3K1_3_equals_0<=TAOK_equals_1]]] & [[1<=TAK1_equals_1 | Proliferation_equals_1<=1] & 1<=p70_equals_1]]]]] | ~ [CREB_equals_1<=PKC_equals_0]] | [FOS_equals_1<=0 | 1<=RSK_equals_0]]]
normalized: ~ [E [true U ~ [[[FOS_equals_1<=0 | 1<=RSK_equals_0] | [~ [CREB_equals_1<=PKC_equals_0] | [[~ [[[[1<=TAK1_equals_1 | Proliferation_equals_1<=1] & 1<=p70_equals_1] & [~ [[1<=MEK1_2_equals_1 & MAP3K1_3_equals_0<=TAOK_equals_1]] & [~ [TGFBR_stimulus_equals_0<=1] | [MAP3K1_3_equals_1<=FGFR3_stimulus_equals_0 | 1<=ATM_equals_1]]]]] | [[MYC_equals_1<=Growth_Arrest_equals_1 | 1<=SPRY_equals_0] & [~ [1<=Growth_Arrest_equals_1] & 1<=Apoptosis_equals_0]]] & ~ [MTK1_equals_1<=DUSP1_equals_1]]]]]]]
abstracting: (MTK1_equals_1<=DUSP1_equals_1)
states: 6,029,313 (6)
abstracting: (1<=Apoptosis_equals_0)
states: 4,063,233 (6)
abstracting: (1<=Growth_Arrest_equals_1)
states: 4,063,232 (6)
abstracting: (1<=SPRY_equals_0)
states: 8,126,465 (6)
abstracting: (MYC_equals_1<=Growth_Arrest_equals_1)
states: 6,094,849 (6)
abstracting: (1<=ATM_equals_1)
states: 8,126,464 (6)
abstracting: (MAP3K1_3_equals_1<=FGFR3_stimulus_equals_0)
states: 8,126,465 (6)
abstracting: (TGFBR_stimulus_equals_0<=1)
states: 8,126,465 (6)
abstracting: (MAP3K1_3_equals_0<=TAOK_equals_1)
states: 4,194,304 (6)
abstracting: (1<=MEK1_2_equals_1)
states: 0
abstracting: (1<=p70_equals_1)
states: 0
abstracting: (Proliferation_equals_1<=1)
states: 8,126,465 (6)
abstracting: (1<=TAK1_equals_1)
states: 0
abstracting: (CREB_equals_1<=PKC_equals_0)
states: 8,126,465 (6)
abstracting: (1<=RSK_equals_0)
states: 8,126,465 (6)
abstracting: (FOS_equals_1<=0)
states: 8,126,465 (6)
-> the formula is TRUE
FORMULA MAPKbis-PT-5320-ReachabilityCardinality-13 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.003sec
checking: EF [[~ [[[[[[[[1<=PDK1_equals_1 | FGFR3_equals_0<=PTEN_equals_1] & [1<=CREB_equals_0 | 1<=p53_equals_0]] & [~ [1<=DNA_damage_equals_0] | [1<=MTK1_equals_0 & AKT_equals_1<=0]]] & BCL2_equals_0<=GAB1_equals_0] & ~ [PTEN_equals_0<=AKT_equals_1]] & ~ [MDM2_equals_0<=TAK1_equals_1]] & ~ [[[~ [[[1<=p14_equals_1 & TGFBR_equals_1<=1] & ~ [RAS_equals_0<=Proliferation_equals_1]]] | [[1<=GRB2_equals_0 & ~ [Proliferation_equals_0<=0]] & [1<=AP1_equals_0 & ~ [1<=MSK_equals_0]]]] & FGFR3_equals_0<=1]]]] & ~ [p53_equals_1<=TGFBR_equals_0]]]
normalized: E [true U [~ [[~ [[[[[~ [1<=MSK_equals_0] & 1<=AP1_equals_0] & [~ [Proliferation_equals_0<=0] & 1<=GRB2_equals_0]] | ~ [[~ [RAS_equals_0<=Proliferation_equals_1] & [1<=p14_equals_1 & TGFBR_equals_1<=1]]]] & FGFR3_equals_0<=1]] & [[[[[[1<=CREB_equals_0 | 1<=p53_equals_0] & [1<=PDK1_equals_1 | FGFR3_equals_0<=PTEN_equals_1]] & [[1<=MTK1_equals_0 & AKT_equals_1<=0] | ~ [1<=DNA_damage_equals_0]]] & BCL2_equals_0<=GAB1_equals_0] & ~ [PTEN_equals_0<=AKT_equals_1]] & ~ [MDM2_equals_0<=TAK1_equals_1]]]] & ~ [p53_equals_1<=TGFBR_equals_0]]]
abstracting: (p53_equals_1<=TGFBR_equals_0)
states: 8,126,465 (6)
abstracting: (MDM2_equals_0<=TAK1_equals_1)
states: 4,063,232 (6)
abstracting: (PTEN_equals_0<=AKT_equals_1)
states: 4,063,232 (6)
abstracting: (BCL2_equals_0<=GAB1_equals_0)
states: 8,126,465 (6)
abstracting: (1<=DNA_damage_equals_0)
states: 0
abstracting: (AKT_equals_1<=0)
states: 8,126,465 (6)
abstracting: (1<=MTK1_equals_0)
states: 3,932,161 (6)
abstracting: (FGFR3_equals_0<=PTEN_equals_1)
states: 4,063,232 (6)
abstracting: (1<=PDK1_equals_1)
states: 0
abstracting: (1<=p53_equals_0)
states: 3,932,161 (6)
abstracting: (1<=CREB_equals_0)
states: 4,063,233 (6)
abstracting: (FGFR3_equals_0<=1)
states: 8,126,465 (6)
abstracting: (TGFBR_equals_1<=1)
states: 8,126,465 (6)
abstracting: (1<=p14_equals_1)
states: 4,063,232 (6)
abstracting: (RAS_equals_0<=Proliferation_equals_1)
states: 0
abstracting: (1<=GRB2_equals_0)
states: 8,126,465 (6)
abstracting: (Proliferation_equals_0<=0)
states: 0
abstracting: (1<=AP1_equals_0)
states: 4,063,233 (6)
abstracting: (1<=MSK_equals_0)
states: 4,063,233 (6)
-> the formula is FALSE
FORMULA MAPKbis-PT-5320-ReachabilityCardinality-07 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.004sec
checking: AG [[[RSK_equals_0<=ELK1_equals_1 | [[[FRS2_equals_1<=0 | [[~ [[JNK_equals_1<=RSK_equals_0 & ATM_equals_0<=Apoptosis_equals_0]] & ~ [[ELK1_equals_1<=PPP2CA_equals_0 | p38_equals_0<=0]]] | [~ [MAX_equals_0<=DUSP1_equals_1] | [[FGFR3_equals_0<=0 | MYC_equals_0<=0] & [1<=DUSP1_equals_1 & 1<=EGFR_equals_0]]]]] | [~ [[~ [FRS2_equals_1<=GRB2_equals_0] & [[AP1_equals_0<=MAX_equals_1 & FOXO3_equals_0<=JNK_equals_0] | [ERK_equals_0<=MEK1_2_equals_0 & ATF2_equals_0<=1]]]] & 1<=FRS2_equals_0]] | [ATM_equals_1<=0 & 1<=PI3K_equals_1]]] | FRS2_equals_1<=DNA_damage_equals_0]]
normalized: ~ [E [true U ~ [[FRS2_equals_1<=DNA_damage_equals_0 | [RSK_equals_0<=ELK1_equals_1 | [[ATM_equals_1<=0 & 1<=PI3K_equals_1] | [[FRS2_equals_1<=0 | [[~ [[ELK1_equals_1<=PPP2CA_equals_0 | p38_equals_0<=0]] & ~ [[JNK_equals_1<=RSK_equals_0 & ATM_equals_0<=Apoptosis_equals_0]]] | [~ [MAX_equals_0<=DUSP1_equals_1] | [[1<=DUSP1_equals_1 & 1<=EGFR_equals_0] & [FGFR3_equals_0<=0 | MYC_equals_0<=0]]]]] | [1<=FRS2_equals_0 & ~ [[~ [FRS2_equals_1<=GRB2_equals_0] & [[ERK_equals_0<=MEK1_2_equals_0 & ATF2_equals_0<=1] | [AP1_equals_0<=MAX_equals_1 & FOXO3_equals_0<=JNK_equals_0]]]]]]]]]]]]
abstracting: (FOXO3_equals_0<=JNK_equals_0)
states: 6,094,849 (6)
abstracting: (AP1_equals_0<=MAX_equals_1)
states: 6,094,848 (6)
abstracting: (ATF2_equals_0<=1)
states: 8,126,465 (6)
abstracting: (ERK_equals_0<=MEK1_2_equals_0)
states: 8,126,465 (6)
abstracting: (FRS2_equals_1<=GRB2_equals_0)
states: 8,126,465 (6)
abstracting: (1<=FRS2_equals_0)
states: 8,126,465 (6)
abstracting: (MYC_equals_0<=0)
states: 4,063,232 (6)
abstracting: (FGFR3_equals_0<=0)
states: 0
abstracting: (1<=EGFR_equals_0)
states: 8,126,465 (6)
abstracting: (1<=DUSP1_equals_1)
states: 4,063,232 (6)
abstracting: (MAX_equals_0<=DUSP1_equals_1)
states: 6,094,848 (6)
abstracting: (ATM_equals_0<=Apoptosis_equals_0)
states: 8,126,465 (6)
abstracting: (JNK_equals_1<=RSK_equals_0)
states: 8,126,465 (6)
abstracting: (p38_equals_0<=0)
states: 3,932,160 (6)
abstracting: (ELK1_equals_1<=PPP2CA_equals_0)
states: 6,094,849 (6)
abstracting: (FRS2_equals_1<=0)
states: 8,126,465 (6)
abstracting: (1<=PI3K_equals_1)
states: 0
abstracting: (ATM_equals_1<=0)
states: 1
abstracting: (RSK_equals_0<=ELK1_equals_1)
states: 4,063,232 (6)
abstracting: (FRS2_equals_1<=DNA_damage_equals_0)
states: 8,126,465 (6)
-> the formula is TRUE
FORMULA MAPKbis-PT-5320-ReachabilityCardinality-04 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.004sec
checking: AG [[AKT_0<=FRS2_equals_0 | [[[[1<=JUN_equals_0 & [~ [[~ [FRS2_equals_1<=ATM_equals_0] & [MDM2_equals_0<=JNK_equals_0 | FGFR3_equals_0<=TGFBR_stimulus_equals_1]]] & 1<=MEK1_2_equals_0]] | ~ [RSK_equals_1<=0]] | [[[AKT_0<=1 & ~ [[[Proliferation_equals_0<=0 | Proliferation_equals_1<=ATM_equals_1] & ~ [TGFBR_equals_1<=RAS_equals_1]]]] | [~ [1<=ELK1_equals_0] & [[[Growth_Arrest_equals_1<=p70_equals_0 | p14_equals_0<=GRB2_equals_1] & p70_equals_1<=EGFR_equals_0] | [PDK1_equals_0<=1 & [FGFR3_stimulus_equals_0<=ELK1_equals_0 & SPRY_equals_1<=DUSP1_equals_1]]]]] | [~ [[[PPP2CA_equals_1<=RAF_equals_0 | 1<=ERK_equals_0] & ~ [MYC_equals_0<=FRS2_equals_0]]] & [1<=CREB_equals_1 & ~ [[1<=FOXO3_equals_1 | [BCL2_equals_0<=0 | 1<=Growth_Arrest_equals_1]]]]]]] | ~ [[~ [Proliferation_equals_0<=PPP2CA_equals_1] & 1<=AKT_equals_1]]]]]
normalized: ~ [E [true U ~ [[AKT_0<=FRS2_equals_0 | [~ [[1<=AKT_equals_1 & ~ [Proliferation_equals_0<=PPP2CA_equals_1]]] | [[[[1<=CREB_equals_1 & ~ [[1<=FOXO3_equals_1 | [BCL2_equals_0<=0 | 1<=Growth_Arrest_equals_1]]]] & ~ [[~ [MYC_equals_0<=FRS2_equals_0] & [PPP2CA_equals_1<=RAF_equals_0 | 1<=ERK_equals_0]]]] | [[[[PDK1_equals_0<=1 & [FGFR3_stimulus_equals_0<=ELK1_equals_0 & SPRY_equals_1<=DUSP1_equals_1]] | [p70_equals_1<=EGFR_equals_0 & [Growth_Arrest_equals_1<=p70_equals_0 | p14_equals_0<=GRB2_equals_1]]] & ~ [1<=ELK1_equals_0]] | [AKT_0<=1 & ~ [[~ [TGFBR_equals_1<=RAS_equals_1] & [Proliferation_equals_0<=0 | Proliferation_equals_1<=ATM_equals_1]]]]]] | [~ [RSK_equals_1<=0] | [1<=JUN_equals_0 & [1<=MEK1_2_equals_0 & ~ [[[MDM2_equals_0<=JNK_equals_0 | FGFR3_equals_0<=TGFBR_stimulus_equals_1] & ~ [FRS2_equals_1<=ATM_equals_0]]]]]]]]]]]]
abstracting: (FRS2_equals_1<=ATM_equals_0)
states: 8,126,465 (6)
abstracting: (FGFR3_equals_0<=TGFBR_stimulus_equals_1)
states: 0
abstracting: (MDM2_equals_0<=JNK_equals_0)
states: 6,094,849 (6)
abstracting: (1<=MEK1_2_equals_0)
states: 8,126,465 (6)
abstracting: (1<=JUN_equals_0)
states: 4,063,233 (6)
abstracting: (RSK_equals_1<=0)
states: 8,126,465 (6)
abstracting: (Proliferation_equals_1<=ATM_equals_1)
states: 8,126,465 (6)
abstracting: (Proliferation_equals_0<=0)
states: 0
abstracting: (TGFBR_equals_1<=RAS_equals_1)
states: 8,126,465 (6)
abstracting: (AKT_0<=1)
states: 8,126,465 (6)
abstracting: (1<=ELK1_equals_0)
states: 4,063,233 (6)
abstracting: (p14_equals_0<=GRB2_equals_1)
states: 4,063,232 (6)
abstracting: (Growth_Arrest_equals_1<=p70_equals_0)
states: 8,126,465 (6)
abstracting: (p70_equals_1<=EGFR_equals_0)
states: 8,126,465 (6)
abstracting: (SPRY_equals_1<=DUSP1_equals_1)
states: 8,126,465 (6)
abstracting: (FGFR3_stimulus_equals_0<=ELK1_equals_0)
states: 4,063,233 (6)
abstracting: (PDK1_equals_0<=1)
states: 8,126,465 (6)
abstracting: (1<=ERK_equals_0)
states: 8,126,465 (6)
abstracting: (PPP2CA_equals_1<=RAF_equals_0)
states: 8,126,465 (6)
abstracting: (MYC_equals_0<=FRS2_equals_0)
states: 8,126,465 (6)
abstracting: (1<=Growth_Arrest_equals_1)
states: 4,063,232 (6)
abstracting: (BCL2_equals_0<=0)
states: 0
abstracting: (1<=FOXO3_equals_1)
states: 4,063,232 (6)
abstracting: (1<=CREB_equals_1)
states: 4,063,232 (6)
abstracting: (Proliferation_equals_0<=PPP2CA_equals_1)
states: 4,063,232 (6)
abstracting: (1<=AKT_equals_1)
states: 0
abstracting: (AKT_0<=FRS2_equals_0)
states: 8,126,465 (6)
-> the formula is TRUE
FORMULA MAPKbis-PT-5320-ReachabilityCardinality-01 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.005sec
checking: EF [~ [[[TGFBR_stimulus_equals_0<=p21_equals_1 | [[[~ [SOS_equals_1<=1] & Proliferation_equals_1<=JUN_equals_1] | PLCG_equals_1<=1] | MAX_equals_1<=0]] | [GRB2_equals_0<=p38_equals_0 & [[~ [[~ [FGFR3_equals_1<=FOS_equals_0] & [[Proliferation_equals_0<=DUSP1_equals_1 & MAP3K1_3_equals_0<=SMAD_equals_0] | ~ [1<=TAK1_equals_1]]]] & [[[[FOS_equals_0<=1 | MEK1_2_equals_1<=SPRY_equals_1] | SPRY_equals_1<=AP1_equals_0] | [~ [1<=SPRY_equals_1] & [RAF_equals_1<=FOS_equals_1 & JNK_equals_0<=0]]] & [[~ [1<=ATM_equals_1] & [TAOK_equals_1<=RSK_equals_1 & ATM_equals_0<=0]] | [~ [MDM2_equals_0<=1] & [p14_equals_1<=EGFR_stimulus_equals_0 & 1<=p53_equals_0]]]]] | [[[~ [[FOS_equals_1<=1 | FGFR3_stimulus_equals_0<=PDK1_equals_1]] & ~ [SPRY_equals_0<=0]] & ~ [[~ [FOXO3_equals_1<=MSK_equals_1] | Proliferation_equals_0<=0]]] & [SPRY_equals_1<=AP1_equals_0 | [DUSP1_equals_0<=MSK_equals_1 | [~ [PDK1_equals_1<=1] & ~ [1<=GRB2_equals_1]]]]]]]]]]
normalized: E [true U ~ [[[GRB2_equals_0<=p38_equals_0 & [[[SPRY_equals_1<=AP1_equals_0 | [DUSP1_equals_0<=MSK_equals_1 | [~ [1<=GRB2_equals_1] & ~ [PDK1_equals_1<=1]]]] & [~ [[Proliferation_equals_0<=0 | ~ [FOXO3_equals_1<=MSK_equals_1]]] & [~ [SPRY_equals_0<=0] & ~ [[FOS_equals_1<=1 | FGFR3_stimulus_equals_0<=PDK1_equals_1]]]]] | [[[[[p14_equals_1<=EGFR_stimulus_equals_0 & 1<=p53_equals_0] & ~ [MDM2_equals_0<=1]] | [[TAOK_equals_1<=RSK_equals_1 & ATM_equals_0<=0] & ~ [1<=ATM_equals_1]]] & [[[RAF_equals_1<=FOS_equals_1 & JNK_equals_0<=0] & ~ [1<=SPRY_equals_1]] | [SPRY_equals_1<=AP1_equals_0 | [FOS_equals_0<=1 | MEK1_2_equals_1<=SPRY_equals_1]]]] & ~ [[[~ [1<=TAK1_equals_1] | [Proliferation_equals_0<=DUSP1_equals_1 & MAP3K1_3_equals_0<=SMAD_equals_0]] & ~ [FGFR3_equals_1<=FOS_equals_0]]]]]] | [TGFBR_stimulus_equals_0<=p21_equals_1 | [MAX_equals_1<=0 | [PLCG_equals_1<=1 | [Proliferation_equals_1<=JUN_equals_1 & ~ [SOS_equals_1<=1]]]]]]]]
abstracting: (SOS_equals_1<=1)
states: 8,126,465 (6)
abstracting: (Proliferation_equals_1<=JUN_equals_1)
states: 8,126,465 (6)
abstracting: (PLCG_equals_1<=1)
states: 8,126,465 (6)
abstracting: (MAX_equals_1<=0)
states: 4,063,233 (6)
abstracting: (TGFBR_stimulus_equals_0<=p21_equals_1)
states: 4,063,232 (6)
abstracting: (FGFR3_equals_1<=FOS_equals_0)
states: 8,126,465 (6)
abstracting: (MAP3K1_3_equals_0<=SMAD_equals_0)
states: 8,126,465 (6)
abstracting: (Proliferation_equals_0<=DUSP1_equals_1)
states: 4,063,232 (6)
abstracting: (1<=TAK1_equals_1)
states: 0
abstracting: (MEK1_2_equals_1<=SPRY_equals_1)
states: 8,126,465 (6)
abstracting: (FOS_equals_0<=1)
states: 8,126,465 (6)
abstracting: (SPRY_equals_1<=AP1_equals_0)
states: 8,126,465 (6)
abstracting: (1<=SPRY_equals_1)
states: 0
abstracting: (JNK_equals_0<=0)
states: 4,063,232 (6)
abstracting: (RAF_equals_1<=FOS_equals_1)
states: 8,126,465 (6)
abstracting: (1<=ATM_equals_1)
states: 8,126,464 (6)
abstracting: (ATM_equals_0<=0)
states: 8,126,464 (6)
abstracting: (TAOK_equals_1<=RSK_equals_1)
states: 3,932,161 (6)
abstracting: (MDM2_equals_0<=1)
states: 8,126,465 (6)
abstracting: (1<=p53_equals_0)
states: 3,932,161 (6)
abstracting: (p14_equals_1<=EGFR_stimulus_equals_0)
states: 8,126,465 (6)
abstracting: (FGFR3_stimulus_equals_0<=PDK1_equals_1)
states: 0
abstracting: (FOS_equals_1<=1)
states: 8,126,465 (6)
abstracting: (SPRY_equals_0<=0)
states: 0
abstracting: (FOXO3_equals_1<=MSK_equals_1)
states: 6,094,849 (6)
abstracting: (Proliferation_equals_0<=0)
states: 0
abstracting: (PDK1_equals_1<=1)
states: 8,126,465 (6)
abstracting: (1<=GRB2_equals_1)
states: 0
abstracting: (DUSP1_equals_0<=MSK_equals_1)
states: 6,094,848 (6)
abstracting: (SPRY_equals_1<=AP1_equals_0)
states: 8,126,465 (6)
abstracting: (GRB2_equals_0<=p38_equals_0)
states: 4,194,305 (6)
-> the formula is FALSE
FORMULA MAPKbis-PT-5320-ReachabilityCardinality-05 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.005sec
checking: EF [[~ [[[[[[~ [1<=PI3K_equals_1] | [TAOK_equals_1<=0 & Growth_Arrest_equals_1<=p38_equals_0]] & [EGFR_stimulus_equals_1<=0 | [MAP3K1_3_equals_0<=MYC_equals_1 & MAX_equals_0<=p70_equals_1]]] | [AKT_0<=0 & [~ [MAP3K1_3_equals_1<=CREB_equals_0] & [CREB_equals_0<=p70_equals_0 | 1<=Apoptosis_equals_1]]]] & ~ [[~ [FOXO3_equals_1<=ELK1_equals_1] & 1<=p70_equals_1]]] | [[[[[1<=DNA_damage_equals_0 | RAF_equals_0<=FGFR3_equals_1] | [PTEN_equals_1<=p53_equals_0 | 1<=MDM2_equals_0]] | ~ [FGFR3_stimulus_equals_1<=RSK_equals_0]] | [1<=SPRY_equals_1 | [[EGFR_equals_1<=1 & PPP2CA_equals_0<=1] & [1<=TGFBR_equals_0 | EGFR_stimulus_equals_0<=FRS2_equals_0]]]] | [~ [[[1<=FRS2_equals_1 & p21_equals_1<=ATF2_equals_1] & [1<=PLCG_equals_1 & ELK1_equals_1<=1]]] & TAK1_equals_1<=MAX_equals_0]]]] & [1<=FGFR3_stimulus_equals_0 & ~ [[BCL2_equals_0<=Growth_Arrest_equals_0 | [MEK1_2_equals_0<=AP1_equals_1 & [1<=PDK1_equals_1 & [1<=PKC_equals_1 & GRB2_equals_0<=p14_equals_1]]]]]]]]
normalized: E [true U [~ [[[~ [[1<=p70_equals_1 & ~ [FOXO3_equals_1<=ELK1_equals_1]]] & [[AKT_0<=0 & [[CREB_equals_0<=p70_equals_0 | 1<=Apoptosis_equals_1] & ~ [MAP3K1_3_equals_1<=CREB_equals_0]]] | [[EGFR_stimulus_equals_1<=0 | [MAP3K1_3_equals_0<=MYC_equals_1 & MAX_equals_0<=p70_equals_1]] & [~ [1<=PI3K_equals_1] | [TAOK_equals_1<=0 & Growth_Arrest_equals_1<=p38_equals_0]]]]] | [[[~ [FGFR3_stimulus_equals_1<=RSK_equals_0] | [[PTEN_equals_1<=p53_equals_0 | 1<=MDM2_equals_0] | [1<=DNA_damage_equals_0 | RAF_equals_0<=FGFR3_equals_1]]] | [1<=SPRY_equals_1 | [[EGFR_equals_1<=1 & PPP2CA_equals_0<=1] & [1<=TGFBR_equals_0 | EGFR_stimulus_equals_0<=FRS2_equals_0]]]] | [TAK1_equals_1<=MAX_equals_0 & ~ [[[1<=FRS2_equals_1 & p21_equals_1<=ATF2_equals_1] & [1<=PLCG_equals_1 & ELK1_equals_1<=1]]]]]]] & [1<=FGFR3_stimulus_equals_0 & ~ [[BCL2_equals_0<=Growth_Arrest_equals_0 | [MEK1_2_equals_0<=AP1_equals_1 & [1<=PDK1_equals_1 & [1<=PKC_equals_1 & GRB2_equals_0<=p14_equals_1]]]]]]]]
abstracting: (GRB2_equals_0<=p14_equals_1)
states: 4,063,232 (6)
abstracting: (1<=PKC_equals_1)
states: 0
abstracting: (1<=PDK1_equals_1)
states: 0
abstracting: (MEK1_2_equals_0<=AP1_equals_1)
states: 4,063,232 (6)
abstracting: (BCL2_equals_0<=Growth_Arrest_equals_0)
states: 4,063,233 (6)
abstracting: (1<=FGFR3_stimulus_equals_0)
states: 8,126,465 (6)
abstracting: (ELK1_equals_1<=1)
states: 8,126,465 (6)
abstracting: (1<=PLCG_equals_1)
states: 0
abstracting: (p21_equals_1<=ATF2_equals_1)
states: 6,094,849 (6)
abstracting: (1<=FRS2_equals_1)
states: 0
abstracting: (TAK1_equals_1<=MAX_equals_0)
states: 8,126,465 (6)
abstracting: (EGFR_stimulus_equals_0<=FRS2_equals_0)
states: 8,126,465 (6)
abstracting: (1<=TGFBR_equals_0)
states: 8,126,465 (6)
abstracting: (PPP2CA_equals_0<=1)
states: 8,126,465 (6)
abstracting: (EGFR_equals_1<=1)
states: 8,126,465 (6)
abstracting: (1<=SPRY_equals_1)
states: 0
abstracting: (RAF_equals_0<=FGFR3_equals_1)
states: 0
abstracting: (1<=DNA_damage_equals_0)
states: 0
abstracting: (1<=MDM2_equals_0)
states: 4,063,233 (6)
abstracting: (PTEN_equals_1<=p53_equals_0)
states: 6,029,313 (6)
abstracting: (FGFR3_stimulus_equals_1<=RSK_equals_0)
states: 8,126,465 (6)
abstracting: (Growth_Arrest_equals_1<=p38_equals_0)
states: 6,160,385 (6)
abstracting: (TAOK_equals_1<=0)
states: 3,932,161 (6)
abstracting: (1<=PI3K_equals_1)
states: 0
abstracting: (MAX_equals_0<=p70_equals_1)
states: 4,063,232 (6)
abstracting: (MAP3K1_3_equals_0<=MYC_equals_1)
states: 4,063,232 (6)
abstracting: (EGFR_stimulus_equals_1<=0)
states: 8,126,465 (6)
abstracting: (MAP3K1_3_equals_1<=CREB_equals_0)
states: 8,126,465 (6)
abstracting: (1<=Apoptosis_equals_1)
states: 4,063,232 (6)
abstracting: (CREB_equals_0<=p70_equals_0)
states: 8,126,465 (6)
abstracting: (AKT_0<=0)
states: 0
abstracting: (FOXO3_equals_1<=ELK1_equals_1)
states: 6,094,849 (6)
abstracting: (1<=p70_equals_1)
states: 0
-> the formula is FALSE
FORMULA MAPKbis-PT-5320-ReachabilityCardinality-00 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.006sec
checking: AG [[[PPP2CA_equals_1<=1 | [~ [MYC_equals_1<=1] & [EGFR_equals_1<=SMAD_equals_1 | ~ [RAF_equals_0<=0]]]] | [[Growth_Arrest_equals_0<=ERK_equals_1 & [[~ [[[[1<=FGFR3_stimulus_equals_0 & SOS_equals_0<=RSK_equals_1] | ~ [1<=PI3K_equals_1]] | [1<=AKT_equals_1 & MSK_equals_0<=MAP3K1_3_equals_1]]] | [[[[Apoptosis_equals_0<=p14_equals_1 & TAK1_equals_0<=1] & [GADD45_equals_0<=MSK_equals_1 & p70_equals_0<=0]] & MAX_equals_0<=0] & [MYC_equals_1<=1 | [[SPRY_equals_0<=1 & Growth_Arrest_equals_1<=ERK_equals_0] | ~ [GADD45_equals_1<=1]]]]] & ~ [[~ [SPRY_equals_1<=AP1_equals_0] & ~ [[[FGFR3_stimulus_equals_0<=0 | 1<=FGFR3_stimulus_equals_0] | ~ [ATM_equals_0<=1]]]]]]] & [1<=TGFBR_equals_1 & [[[[~ [1<=ELK1_equals_1] | ~ [[1<=FOS_equals_0 & Apoptosis_equals_0<=p21_equals_1]]] | [~ [[RAS_equals_0<=TAK1_equals_0 & FOS_equals_1<=1]] & [[TAOK_equals_1<=GAB1_equals_0 & ATF2_equals_1<=0] | [1<=CREB_equals_0 & SOS_equals_0<=EGFR_stimulus_equals_0]]]] & [JNK_equals_1<=0 & ~ [[1<=DUSP1_equals_0 | ATM_equals_0<=1]]]] & [RSK_equals_0<=0 | PLCG_equals_1<=JUN_equals_1]]]]]]
normalized: ~ [E [true U ~ [[[[1<=TGFBR_equals_1 & [[RSK_equals_0<=0 | PLCG_equals_1<=JUN_equals_1] & [[JNK_equals_1<=0 & ~ [[1<=DUSP1_equals_0 | ATM_equals_0<=1]]] & [[[[1<=CREB_equals_0 & SOS_equals_0<=EGFR_stimulus_equals_0] | [TAOK_equals_1<=GAB1_equals_0 & ATF2_equals_1<=0]] & ~ [[RAS_equals_0<=TAK1_equals_0 & FOS_equals_1<=1]]] | [~ [[1<=FOS_equals_0 & Apoptosis_equals_0<=p21_equals_1]] | ~ [1<=ELK1_equals_1]]]]]] & [Growth_Arrest_equals_0<=ERK_equals_1 & [~ [[~ [[~ [ATM_equals_0<=1] | [FGFR3_stimulus_equals_0<=0 | 1<=FGFR3_stimulus_equals_0]]] & ~ [SPRY_equals_1<=AP1_equals_0]]] & [[[MYC_equals_1<=1 | [~ [GADD45_equals_1<=1] | [SPRY_equals_0<=1 & Growth_Arrest_equals_1<=ERK_equals_0]]] & [MAX_equals_0<=0 & [[GADD45_equals_0<=MSK_equals_1 & p70_equals_0<=0] & [Apoptosis_equals_0<=p14_equals_1 & TAK1_equals_0<=1]]]] | ~ [[[1<=AKT_equals_1 & MSK_equals_0<=MAP3K1_3_equals_1] | [~ [1<=PI3K_equals_1] | [1<=FGFR3_stimulus_equals_0 & SOS_equals_0<=RSK_equals_1]]]]]]]] | [PPP2CA_equals_1<=1 | [[EGFR_equals_1<=SMAD_equals_1 | ~ [RAF_equals_0<=0]] & ~ [MYC_equals_1<=1]]]]]]]
abstracting: (MYC_equals_1<=1)
states: 8,126,465 (6)
abstracting: (RAF_equals_0<=0)
states: 0
abstracting: (EGFR_equals_1<=SMAD_equals_1)
states: 8,126,465 (6)
abstracting: (PPP2CA_equals_1<=1)
states: 8,126,465 (6)
abstracting: (SOS_equals_0<=RSK_equals_1)
states: 0
abstracting: (1<=FGFR3_stimulus_equals_0)
states: 8,126,465 (6)
abstracting: (1<=PI3K_equals_1)
states: 0
abstracting: (MSK_equals_0<=MAP3K1_3_equals_1)
states: 4,063,232 (6)
abstracting: (1<=AKT_equals_1)
states: 0
abstracting: (TAK1_equals_0<=1)
states: 8,126,465 (6)
abstracting: (Apoptosis_equals_0<=p14_equals_1)
states: 6,094,848 (6)
abstracting: (p70_equals_0<=0)
states: 0
abstracting: (GADD45_equals_0<=MSK_equals_1)
states: 6,029,312 (6)
abstracting: (MAX_equals_0<=0)
states: 4,063,232 (6)
abstracting: (Growth_Arrest_equals_1<=ERK_equals_0)
states: 8,126,465 (6)
abstracting: (SPRY_equals_0<=1)
states: 8,126,465 (6)
abstracting: (GADD45_equals_1<=1)
states: 8,126,465 (6)
abstracting: (MYC_equals_1<=1)
states: 8,126,465 (6)
abstracting: (SPRY_equals_1<=AP1_equals_0)
states: 8,126,465 (6)
abstracting: (1<=FGFR3_stimulus_equals_0)
states: 8,126,465 (6)
abstracting: (FGFR3_stimulus_equals_0<=0)
states: 0
abstracting: (ATM_equals_0<=1)
states: 8,126,465 (6)
abstracting: (Growth_Arrest_equals_0<=ERK_equals_1)
states: 4,063,232 (6)
abstracting: (1<=ELK1_equals_1)
states: 4,063,232 (6)
abstracting: (Apoptosis_equals_0<=p21_equals_1)
states: 6,094,848 (6)
abstracting: (1<=FOS_equals_0)
states: 8,126,465 (6)
abstracting: (FOS_equals_1<=1)
states: 8,126,465 (6)
abstracting: (RAS_equals_0<=TAK1_equals_0)
states: 8,126,465 (6)
abstracting: (ATF2_equals_1<=0)
states: 4,063,233 (6)
abstracting: (TAOK_equals_1<=GAB1_equals_0)
states: 8,126,465 (6)
abstracting: (SOS_equals_0<=EGFR_stimulus_equals_0)
states: 8,126,465 (6)
abstracting: (1<=CREB_equals_0)
states: 4,063,233 (6)
abstracting: (ATM_equals_0<=1)
states: 8,126,465 (6)
abstracting: (1<=DUSP1_equals_0)
states: 4,063,233 (6)
abstracting: (JNK_equals_1<=0)
states: 4,063,233 (6)
abstracting: (PLCG_equals_1<=JUN_equals_1)
states: 8,126,465 (6)
abstracting: (RSK_equals_0<=0)
states: 0
abstracting: (1<=TGFBR_equals_1)
states: 0
-> the formula is TRUE
FORMULA MAPKbis-PT-5320-ReachabilityCardinality-12 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.005sec
checking: AG [[[~ [RAS_equals_1<=0] | ~ [[[[MYC_equals_1<=FGFR3_stimulus_equals_0 | [MEK1_2_equals_1<=0 | [[FGFR3_equals_0<=1 & 1<=MEK1_2_equals_0] | 1<=BCL2_equals_0]]] & [PKC_equals_1<=1 & 1<=JUN_equals_1]] & MSK_equals_1<=p38_equals_1]]] | [[[p21_equals_0<=Apoptosis_equals_1 & GADD45_equals_1<=0] & [[MYC_equals_0<=0 | [[~ [[EGFR_equals_1<=1 & p38_equals_1<=FGFR3_equals_1]] | ~ [[TAOK_equals_1<=TGFBR_stimulus_equals_1 | MAP3K1_3_equals_0<=0]]] & [1<=PKC_equals_0 & [~ [FRS2_equals_1<=1] & [1<=FOXO3_equals_1 | 1<=SOS_equals_0]]]]] & [[1<=p70_equals_1 | ~ [p21_equals_0<=PPP2CA_equals_0]] & [1<=ATM_equals_1 | 1<=TGFBR_stimulus_equals_0]]]] | [[[[~ [ATF2_equals_0<=TGFBR_equals_0] & ~ [[MDM2_equals_1<=1 | 1<=JNK_equals_0]]] | ~ [[~ [PDK1_equals_0<=1] & ~ [1<=p53_equals_1]]]] | [[[~ [[1<=GRB2_equals_0 | ELK1_equals_0<=FOS_equals_1]] | [[PI3K_equals_1<=0 | p53_equals_1<=RAS_equals_0] | JUN_equals_1<=1]] | [[[1<=p38_equals_0 & 1<=PLCG_equals_1] | ~ [RSK_equals_1<=1]] & [[FGFR3_equals_1<=0 & 1<=GRB2_equals_1] | [EGFR_stimulus_equals_1<=1 & SPRY_equals_0<=JNK_equals_0]]]] | ~ [MDM2_equals_1<=TAK1_equals_1]]] | 1<=FOS_equals_0]]]]
normalized: ~ [E [true U ~ [[[[1<=FOS_equals_0 | [[~ [MDM2_equals_1<=TAK1_equals_1] | [[[[EGFR_stimulus_equals_1<=1 & SPRY_equals_0<=JNK_equals_0] | [FGFR3_equals_1<=0 & 1<=GRB2_equals_1]] & [~ [RSK_equals_1<=1] | [1<=p38_equals_0 & 1<=PLCG_equals_1]]] | [[JUN_equals_1<=1 | [PI3K_equals_1<=0 | p53_equals_1<=RAS_equals_0]] | ~ [[1<=GRB2_equals_0 | ELK1_equals_0<=FOS_equals_1]]]]] | [~ [[~ [1<=p53_equals_1] & ~ [PDK1_equals_0<=1]]] | [~ [[MDM2_equals_1<=1 | 1<=JNK_equals_0]] & ~ [ATF2_equals_0<=TGFBR_equals_0]]]]] | [[[[1<=ATM_equals_1 | 1<=TGFBR_stimulus_equals_0] & [1<=p70_equals_1 | ~ [p21_equals_0<=PPP2CA_equals_0]]] & [MYC_equals_0<=0 | [[1<=PKC_equals_0 & [[1<=FOXO3_equals_1 | 1<=SOS_equals_0] & ~ [FRS2_equals_1<=1]]] & [~ [[TAOK_equals_1<=TGFBR_stimulus_equals_1 | MAP3K1_3_equals_0<=0]] | ~ [[EGFR_equals_1<=1 & p38_equals_1<=FGFR3_equals_1]]]]]] & [p21_equals_0<=Apoptosis_equals_1 & GADD45_equals_1<=0]]] | [~ [[MSK_equals_1<=p38_equals_1 & [[PKC_equals_1<=1 & 1<=JUN_equals_1] & [MYC_equals_1<=FGFR3_stimulus_equals_0 | [MEK1_2_equals_1<=0 | [1<=BCL2_equals_0 | [FGFR3_equals_0<=1 & 1<=MEK1_2_equals_0]]]]]]] | ~ [RAS_equals_1<=0]]]]]]
abstracting: (RAS_equals_1<=0)
states: 8,126,465 (6)
abstracting: (1<=MEK1_2_equals_0)
states: 8,126,465 (6)
abstracting: (FGFR3_equals_0<=1)
states: 8,126,465 (6)
abstracting: (1<=BCL2_equals_0)
states: 8,126,465 (6)
abstracting: (MEK1_2_equals_1<=0)
states: 8,126,465 (6)
abstracting: (MYC_equals_1<=FGFR3_stimulus_equals_0)
states: 8,126,465 (6)
abstracting: (1<=JUN_equals_1)
states: 4,063,232 (6)
abstracting: (PKC_equals_1<=1)
states: 8,126,465 (6)
abstracting: (MSK_equals_1<=p38_equals_1)
states: 6,029,313 (6)
abstracting: (GADD45_equals_1<=0)
states: 4,194,305 (6)
abstracting: (p21_equals_0<=Apoptosis_equals_1)
states: 6,094,848 (6)
abstracting: (p38_equals_1<=FGFR3_equals_1)
states: 4,194,305 (6)
abstracting: (EGFR_equals_1<=1)
states: 8,126,465 (6)
abstracting: (MAP3K1_3_equals_0<=0)
states: 0
abstracting: (TAOK_equals_1<=TGFBR_stimulus_equals_1)
states: 3,932,161 (6)
abstracting: (FRS2_equals_1<=1)
states: 8,126,465 (6)
abstracting: (1<=SOS_equals_0)
states: 8,126,465 (6)
abstracting: (1<=FOXO3_equals_1)
states: 4,063,232 (6)
abstracting: (1<=PKC_equals_0)
states: 8,126,465 (6)
abstracting: (MYC_equals_0<=0)
states: 4,063,232 (6)
abstracting: (p21_equals_0<=PPP2CA_equals_0)
states: 6,094,849 (6)
abstracting: (1<=p70_equals_1)
states: 0
abstracting: (1<=TGFBR_stimulus_equals_0)
states: 8,126,465 (6)
abstracting: (1<=ATM_equals_1)
states: 8,126,464 (6)
abstracting: (ATF2_equals_0<=TGFBR_equals_0)
states: 8,126,465 (6)
abstracting: (1<=JNK_equals_0)
states: 4,063,233 (6)
abstracting: (MDM2_equals_1<=1)
states: 8,126,465 (6)
abstracting: (PDK1_equals_0<=1)
states: 8,126,465 (6)
abstracting: (1<=p53_equals_1)
states: 4,194,304 (6)
abstracting: (ELK1_equals_0<=FOS_equals_1)
states: 4,063,232 (6)
abstracting: (1<=GRB2_equals_0)
states: 8,126,465 (6)
abstracting: (p53_equals_1<=RAS_equals_0)
states: 8,126,465 (6)
abstracting: (PI3K_equals_1<=0)
states: 8,126,465 (6)
abstracting: (JUN_equals_1<=1)
states: 8,126,465 (6)
abstracting: (1<=PLCG_equals_1)
states: 0
abstracting: (1<=p38_equals_0)
states: 4,194,305 (6)
abstracting: (RSK_equals_1<=1)
states: 8,126,465 (6)
abstracting: (1<=GRB2_equals_1)
states: 0
abstracting: (FGFR3_equals_1<=0)
states: 8,126,465 (6)
abstracting: (SPRY_equals_0<=JNK_equals_0)
states: 4,063,233 (6)
abstracting: (EGFR_stimulus_equals_1<=1)
states: 8,126,465 (6)
abstracting: (MDM2_equals_1<=TAK1_equals_1)
states: 4,063,233 (6)
abstracting: (1<=FOS_equals_0)
states: 8,126,465 (6)
-> the formula is TRUE
FORMULA MAPKbis-PT-5320-ReachabilityCardinality-10 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.006sec
totally nodes used: 22838 (2.3e+04)
number of garbage collections: 0
fire ops cache: hits/miss/sum: 41203 154671 195874
used/not used/entry size/cache size: 160919 66947945 16 1024MB
basic ops cache: hits/miss/sum: 12412 53735 66147
used/not used/entry size/cache size: 83316 16693900 12 192MB
unary ops cache: hits/miss/sum: 0 0 0
used/not used/entry size/cache size: 0 16777216 8 128MB
abstract ops cache: hits/miss/sum: 0 0 0
used/not used/entry size/cache size: 0 16777216 12 192MB
state nr cache: hits/miss/sum: 1617 4669 6286
used/not used/entry size/cache size: 4668 8383940 32 256MB
max state cache: hits/miss/sum: 0 0 0
used/not used/entry size/cache size: 0 8388608 32 256MB
uniqueHash elements/entry size/size: 67108864 4 256MB
0 67086200
1 22491
2 172
3 1
4 0
5 0
6 0
7 0
8 0
9 0
>= 10 0
Total processing time: 0m 4.687sec
BK_STOP 1678641249420
--------------------
content from stderr:
check for maximal unmarked siphon
found
The net has a maximal unmarked siphon:
PI3K_equals_1
TAK1_equals_1
MAP3K1_3_equals_1
SPRY_equals_1
TGFBR_equals_1
TGFBR_stimulus_equals_1
p70_equals_1
AKT_equals_1
RAS_equals_1
MEK1_2_equals_1
SOS_equals_1
BCL2_equals_1
SMAD_equals_1
EGFR_stimulus_equals_1
DNA_damage_equals_0
FGFR3_equals_1
GAB1_equals_1
FGFR3_stimulus_equals_1
ERK_equals_1
FRS2_equals_1
EGFR_equals_1
RSK_equals_1
PKC_equals_1
GRB2_equals_1
PDK1_equals_1
PLCG_equals_1
RAF_equals_1
FOS_equals_1
Proliferation_equals_1
The net has transition(s) that can never fire:
AKT_equals_1_to_AKT_equals_0_when_PDK1_equals_0
JNK_equals_0_to_JNK_equals_1_when_DUSP1_equals_1_and_MAP3K1_3_equals_1_and_TAK1_equals_1
JNK_equals_0_to_JNK_equals_1_when_DUSP1_equals_1_and_MAP3K1_3_equals_1_and_TAOK_equals_1
FGFR3_equals_0_to_FGFR3_equals_1_when_FGFR3_stimulus_equals_1_and_GRB2_equals_0_and_PKC_equals_0
GRB2_equals_0_to_GRB2_equals_1_when_FRS2_equals_1
GRB2_equals_0_to_GRB2_equals_1_when_TGFBR_equals_1
GRB2_equals_1_to_GRB2_equals_0_when_EGFR_equals_0_and_FRS2_equals_0_and_TGFBR_equals_0
MYC_equals_0_to_MYC_equals_1_when_AKT_equals_1_and_MSK_equals_1
GAB1_equals_0_to_GAB1_equals_1_when_PI3K_equals_1
JNK_equals_1_to_JNK_equals_0_when_DUSP1_equals_1_and_MAP3K1_3_equals_1_and_MTK1_equals_0_and_TAK1_equals_0_and_TAOK_equals_0
JNK_equals_0_to_JNK_equals_1_when_DUSP1_equals_0_and_MAP3K1_3_equals_1
p38_equals_1_to_p38_equals_0_when_DUSP1_equals_1_and_MAP3K1_3_equals_1_and_MTK1_equals_0_and_TAK1_equals_0_and_TAOK_equals_0
p70_equals_0_to_p70_equals_1_when_ERK_equals_1_and_PDK1_equals_1
p70_equals_1_to_p70_equals_0_when_ERK_equals_0
p70_equals_1_to_p70_equals_0_when_PDK1_equals_0
FOS_equals_1_to_FOS_equals_0_when_CREB_equals_0_and_ELK1_equals_0
SMAD_equals_0_to_SMAD_equals_1_when_TGFBR_equals_1
SMAD_equals_1_to_SMAD_equals_0_when_TGFBR_equals_0
MEK1_2_equals_1_to_MEK1_2_equals_0_when_PPP2CA_equals_1
MSK_equals_0_to_MSK_equals_1_when_ERK_equals_1
MAP3K1_3_equals_0_to_MAP3K1_3_equals_1_when_RAS_equals_1
MAP3K1_3_equals_1_to_MAP3K1_3_equals_0_when_RAS_equals_0
BCL2_equals_1_to_BCL2_equals_0_when_CREB_equals_0
MDM2_equals_0_to_MDM2_equals_1_when_AKT_equals_1_and_p14_equals_0
p38_equals_0_to_p38_equals_1_when_DUSP1_equals_1_and_MAP3K1_3_equals_1_and_MTK1_equals_1
p38_equals_0_to_p38_equals_1_when_DUSP1_equals_1_and_MAP3K1_3_equals_1_and_TAK1_equals_1
p38_equals_0_to_p38_equals_1_when_DUSP1_equals_1_and_MAP3K1_3_equals_1_and_TAOK_equals_1
PI3K_equals_1_to_PI3K_equals_0_when_GAB1_equals_0_and_RAS_equals_0
PI3K_equals_1_to_PI3K_equals_0_when_GAB1_equals_0_and_SOS_equals_0
PKC_equals_0_to_PKC_equals_1_when_PLCG_equals_1
PKC_equals_1_to_PKC_equals_0_when_PLCG_equals_0
PLCG_equals_0_to_PLCG_equals_1_when_EGFR_equals_1
GADD45_equals_0_to_GADD45_equals_1_when_SMAD_equals_1
FRS2_equals_0_to_FRS2_equals_1_when_FGFR3_equals_1_and_GRB2_equals_0_and_SPRY_equals_0
FRS2_equals_1_to_FRS2_equals_0_when_FGFR3_equals_0
JNK_equals_0_to_JNK_equals_1_when_DUSP1_equals_0_and_TAK1_equals_1
ERK_equals_0_to_ERK_equals_1_when_MEK1_2_equals_1
JNK_equals_0_to_JNK_equals_1_when_DUSP1_equals_1_and_MAP3K1_3_equals_1_and_MTK1_equals_1
EGFR_equals_1_to_EGFR_equals_0_when_EGFR_stimulus_equals_0_and_SPRY_equals_0
FGFR3_equals_1_to_FGFR3_equals_0_when_GRB2_equals_1
FGFR3_equals_1_to_FGFR3_equals_0_when_PKC_equals_1
FOS_equals_0_to_FOS_equals_1_when_CREB_equals_0_and_ELK1_equals_1_and_ERK_equals_1_and_RSK_equals_1
ELK1_equals_0_to_ELK1_equals_1_when_ERK_equals_1
FGFR3_equals_1_to_FGFR3_equals_0_when_FGFR3_stimulus_equals_0
FOS_equals_1_to_FOS_equals_0_when_ERK_equals_0
FOS_equals_1_to_FOS_equals_0_when_RSK_equals_0
FOS_equals_0_to_FOS_equals_1_when_CREB_equals_1_and_ERK_equals_1_and_RSK_equals_1
PDK1_equals_0_to_PDK1_equals_1_when_PI3K_equals_1
PDK1_equals_1_to_PDK1_equals_0_when_PI3K_equals_0
PI3K_equals_0_to_PI3K_equals_1_when_GAB1_equals_1
PI3K_equals_0_to_PI3K_equals_1_when_RAS_equals_1_and_SOS_equals_1
MEK1_2_equals_1_to_MEK1_2_equals_0_when_AP1_equals_1
GAB1_equals_1_to_GAB1_equals_0_when_GRB2_equals_0_and_PI3K_equals_0
MEK1_2_equals_0_to_MEK1_2_equals_1_when_AP1_equals_0_and_MAP3K1_3_equals_0_and_PPP2CA_equals_0_and_RAF_equals_1
MEK1_2_equals_0_to_MEK1_2_equals_1_when_AP1_equals_0_and_MAP3K1_3_equals_1_and_PPP2CA_equals_0
MEK1_2_equals_1_to_MEK1_2_equals_0_when_MAP3K1_3_equals_0_and_RAF_equals_0
FRS2_equals_1_to_FRS2_equals_0_when_GRB2_equals_1
FRS2_equals_1_to_FRS2_equals_0_when_SPRY_equals_1
FOXO3_equals_1_to_FOXO3_equals_0_when_AKT_equals_1
Proliferation_equals_1_to_Proliferation_equals_0_when_MYC_equals_0
Proliferation_equals_1_to_Proliferation_equals_0_when_p21_equals_1
Proliferation_equals_1_to_Proliferation_equals_0_when_p70_equals_0
RAF_equals_0_to_RAF_equals_1_when_AKT_equals_0_and_ERK_equals_0_and_PKC_equals_1
RAF_equals_0_to_RAF_equals_1_when_AKT_equals_0_and_ERK_equals_0_and_RAS_equals_1
EGFR_equals_1_to_EGFR_equals_0_when_GRB2_equals_1
EGFR_equals_1_to_EGFR_equals_0_when_PKC_equals_1
EGFR_equals_0_to_EGFR_equals_1_when_EGFR_stimulus_equals_0_and_GRB2_equals_0_and_PKC_equals_0_and_SPRY_equals_1
Apoptosis_equals_1_to_Apoptosis_equals_0_when_ERK_equals_1
RSK_equals_1_to_RSK_equals_0_when_ERK_equals_0
JNK_equals_0_to_JNK_equals_1_when_DUSP1_equals_1_and_MAP3K1_3_equals_0_and_MTK1_equals_0_and_TAK1_equals_1_and_TAOK_equals_1
JNK_equals_0_to_JNK_equals_1_when_DUSP1_equals_1_and_MAP3K1_3_equals_0_and_MTK1_equals_1_and_TAK1_equals_1
SOS_equals_0_to_SOS_equals_1_when_GRB2_equals_1_and_RSK_equals_0
SOS_equals_1_to_SOS_equals_0_when_GRB2_equals_0
SOS_equals_1_to_SOS_equals_0_when_RSK_equals_1
SPRY_equals_0_to_SPRY_equals_1_when_ERK_equals_1
RAF_equals_1_to_RAF_equals_0_when_AKT_equals_1
RAF_equals_1_to_RAF_equals_0_when_ERK_equals_1
RAF_equals_1_to_RAF_equals_0_when_PKC_equals_0_and_RAS_equals_0
RAS_equals_0_to_RAS_equals_1_when_PLCG_equals_1
RAS_equals_0_to_RAS_equals_1_when_SOS_equals_1
RAS_equals_1_to_RAS_equals_0_when_PLCG_equals_0_and_SOS_equals_0
RSK_equals_0_to_RSK_equals_1_when_ERK_equals_1
ERK_equals_1_to_ERK_equals_0_when_MEK1_2_equals_0
PLCG_equals_0_to_PLCG_equals_1_when_FGFR3_equals_1
PLCG_equals_1_to_PLCG_equals_0_when_EGFR_equals_0_and_FGFR3_equals_0
Proliferation_equals_0_to_Proliferation_equals_1_when_MYC_equals_1_and_p21_equals_0_and_p70_equals_1
GRB2_equals_0_to_GRB2_equals_1_when_EGFR_equals_1
SPRY_equals_1_to_SPRY_equals_0_when_ERK_equals_0
TAK1_equals_0_to_TAK1_equals_1_when_TGFBR_equals_1
TAK1_equals_1_to_TAK1_equals_0_when_TGFBR_equals_0
TGFBR_equals_0_to_TGFBR_equals_1_when_TGFBR_stimulus_equals_1
TGFBR_equals_1_to_TGFBR_equals_0_when_TGFBR_stimulus_equals_0
p21_equals_1_to_p21_equals_0_when_AKT_equals_1
p38_equals_0_to_p38_equals_1_when_DUSP1_equals_0_and_MAP3K1_3_equals_1
p38_equals_0_to_p38_equals_1_when_DUSP1_equals_0_and_TAK1_equals_1
p38_equals_0_to_p38_equals_1_when_DUSP1_equals_1_and_MAP3K1_3_equals_0_and_MTK1_equals_0_and_TAK1_equals_1_and_TAOK_equals_1
p38_equals_0_to_p38_equals_1_when_DUSP1_equals_1_and_MAP3K1_3_equals_0_and_MTK1_equals_1_and_TAK1_equals_1
AP1_equals_0_to_AP1_equals_1_when_ATF2_equals_0_and_FOS_equals_1_and_JUN_equals_1
EGFR_equals_0_to_EGFR_equals_1_when_EGFR_stimulus_equals_1_and_GRB2_equals_0_and_PKC_equals_0
ATM_equals_1_to_ATM_equals_0_when_DNA_damage_equals_0
BCL2_equals_0_to_BCL2_equals_1_when_AKT_equals_1_and_CREB_equals_1
Apoptosis_equals_1_to_Apoptosis_equals_0_when_BCL2_equals_1
GAB1_equals_0_to_GAB1_equals_1_when_GRB2_equals_1
AKT_equals_1_to_AKT_equals_0_when_PTEN_equals_1
AKT_equals_0_to_AKT_equals_1_when_PDK1_equals_1_and_PTEN_equals_0
BCL2_equals_1_to_BCL2_equals_0_when_AKT_equals_0
check for constant places
DNA_damage_equals_0
DNA_damage_equals_1
EGFR_stimulus_equals_0
EGFR_stimulus_equals_1
FGFR3_stimulus_equals_0
FGFR3_stimulus_equals_1
TGFBR_stimulus_equals_0
TGFBR_stimulus_equals_1
found 8 constant places
check if there are places and transitions
ok
check if there are transitions without pre-places
ok
check if at least one transition is enabled in m0
ok
check if there are transitions that can never fire
ok
initing FirstDep: 0m 0.000sec
iterations count:3466 (20), effective:182 (1)
initing FirstDep: 0m 0.000sec
Sequence of Actions to be Executed by the VM
This is useful if one wants to reexecute the tool in the VM from the submitted image disk.
set -x
# this is for BenchKit: configuration of major elements for the test
export BK_INPUT="MAPKbis-PT-5320"
export BK_EXAMINATION="ReachabilityCardinality"
export BK_TOOL="marcie"
export BK_RESULT_DIR="/tmp/BK_RESULTS/OUTPUTS"
export BK_TIME_CONFINEMENT="3600"
export BK_MEMORY_CONFINEMENT="16384"
export BK_BIN_PATH="/home/mcc/BenchKit/bin/"
# this is specific to your benchmark or test
export BIN_DIR="$HOME/BenchKit/bin"
# remove the execution directoty if it exists (to avoid increse of .vmdk images)
if [ -d execution ] ; then
rm -rf execution
fi
# this is for BenchKit: explicit launching of the test
echo "====================================================================="
echo " Generated by BenchKit 2-5348"
echo " Executing tool marcie"
echo " Input is MAPKbis-PT-5320, examination is ReachabilityCardinality"
echo " Time confinement is $BK_TIME_CONFINEMENT seconds"
echo " Memory confinement is 16384 MBytes"
echo " Number of cores is 1"
echo " Run identifier is r225-tall-167856407900646"
echo "====================================================================="
echo
echo "--------------------"
echo "preparation of the directory to be used:"
tar xzf /home/mcc/BenchKit/INPUTS/MAPKbis-PT-5320.tgz
mv MAPKbis-PT-5320 execution
cd execution
if [ "ReachabilityCardinality" = "ReachabilityDeadlock" ] || [ "ReachabilityCardinality" = "UpperBounds" ] || [ "ReachabilityCardinality" = "QuasiLiveness" ] || [ "ReachabilityCardinality" = "StableMarking" ] || [ "ReachabilityCardinality" = "Liveness" ] || [ "ReachabilityCardinality" = "OneSafe" ] || [ "ReachabilityCardinality" = "StateSpace" ]; then
rm -f GenericPropertiesVerdict.xml
fi
pwd
ls -lh
echo
echo "--------------------"
echo "content from stdout:"
echo
echo "=== Data for post analysis generated by BenchKit (invocation template)"
echo
if [ "ReachabilityCardinality" = "UpperBounds" ] ; then
echo "The expected result is a vector of positive values"
echo NUM_VECTOR
elif [ "ReachabilityCardinality" != "StateSpace" ] ; then
echo "The expected result is a vector of booleans"
echo BOOL_VECTOR
else
echo "no data necessary for post analysis"
fi
echo
if [ -f "ReachabilityCardinality.txt" ] ; then
echo "here is the order used to build the result vector(from text file)"
for x in $(grep Property ReachabilityCardinality.txt | cut -d ' ' -f 2 | sort -u) ; do
echo "FORMULA_NAME $x"
done
elif [ -f "ReachabilityCardinality.xml" ] ; then # for cunf (txt files deleted;-)
echo echo "here is the order used to build the result vector(from xml file)"
for x in $(grep '
echo "FORMULA_NAME $x"
done
elif [ "ReachabilityCardinality" = "ReachabilityDeadlock" ] || [ "ReachabilityCardinality" = "QuasiLiveness" ] || [ "ReachabilityCardinality" = "StableMarking" ] || [ "ReachabilityCardinality" = "Liveness" ] || [ "ReachabilityCardinality" = "OneSafe" ] ; then
echo "FORMULA_NAME ReachabilityCardinality"
fi
echo
echo "=== Now, execution of the tool begins"
echo
echo -n "BK_START "
date -u +%s%3N
echo
timeout -s 9 $BK_TIME_CONFINEMENT bash -c "/home/mcc/BenchKit/BenchKit_head.sh 2> STDERR ; echo ; echo -n \"BK_STOP \" ; date -u +%s%3N"
if [ $? -eq 137 ] ; then
echo
echo "BK_TIME_CONFINEMENT_REACHED"
fi
echo
echo "--------------------"
echo "content from stderr:"
echo
cat STDERR ;

