About the Execution of Tapaal for DNAwalker-PT-03track12BlockBoth
| Execution Summary | |||||
| Max Memory Used (MB) |
Time wait (ms) | CPU Usage (ms) | I/O Wait (ms) | Computed Result | Execution Status |
| 69.596 | 27970.00 | 92134.00 | 30.00 | 2 2 2 2 2 2 2 1 2 2 2 2 1 2 2 2 | normal |
Execution Chart
We display below the execution chart for this examination (boot time has been removed).
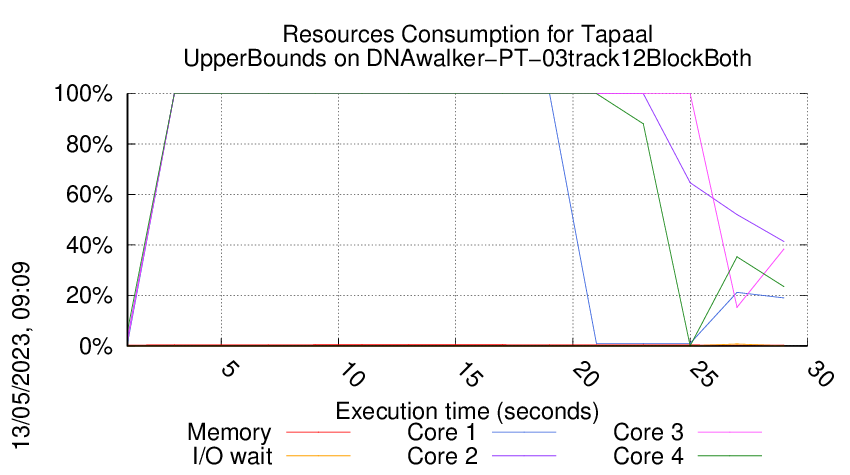
Trace from the execution
Formatting '/data/fkordon/mcc2023-input.r109-tall-167814487700893.qcow2', fmt=qcow2 size=4294967296 backing_file=/data/fkordon/mcc2023-input.qcow2 cluster_size=65536 lazy_refcounts=off refcount_bits=16
Waiting for the VM to be ready (probing ssh)
...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................
=====================================================================
Generated by BenchKit 2-5348
Executing tool tapaal
Input is DNAwalker-PT-03track12BlockBoth, examination is UpperBounds
Time confinement is 3600 seconds
Memory confinement is 16384 MBytes
Number of cores is 4
Run identifier is r109-tall-167814487700893
=====================================================================
--------------------
preparation of the directory to be used:
/home/mcc/execution
total 564K
-rw-r--r-- 1 mcc users 6.8K Feb 25 12:58 CTLCardinality.txt
-rw-r--r-- 1 mcc users 75K Feb 25 12:58 CTLCardinality.xml
-rw-r--r-- 1 mcc users 5.7K Feb 25 12:57 CTLFireability.txt
-rw-r--r-- 1 mcc users 50K Feb 25 12:57 CTLFireability.xml
-rw-r--r-- 1 mcc users 4.2K Jan 29 11:40 GenericPropertiesDefinition.xml
-rw-r--r-- 1 mcc users 3.9K Feb 25 15:54 LTLCardinality.txt
-rw-r--r-- 1 mcc users 26K Feb 25 15:54 LTLCardinality.xml
-rw-r--r-- 1 mcc users 2.4K Feb 25 15:54 LTLFireability.txt
-rw-r--r-- 1 mcc users 17K Feb 25 15:54 LTLFireability.xml
-rw-r--r-- 1 mcc users 15K Feb 25 12:59 ReachabilityCardinality.txt
-rw-r--r-- 1 mcc users 168K Feb 25 12:59 ReachabilityCardinality.xml
-rw-r--r-- 1 mcc users 12K Feb 25 12:59 ReachabilityFireability.txt
-rw-r--r-- 1 mcc users 99K Feb 25 12:59 ReachabilityFireability.xml
-rw-r--r-- 1 mcc users 1.8K Feb 25 15:54 UpperBounds.txt
-rw-r--r-- 1 mcc users 3.9K Feb 25 15:54 UpperBounds.xml
-rw-r--r-- 1 mcc users 6 Mar 5 18:22 equiv_col
-rw-r--r-- 1 mcc users 19 Mar 5 18:22 instance
-rw-r--r-- 1 mcc users 6 Mar 5 18:22 iscolored
-rw-r--r-- 1 mcc users 37K Mar 5 18:22 model.pnml
--------------------
content from stdout:
=== Data for post analysis generated by BenchKit (invocation template)
The expected result is a vector of positive values
NUM_VECTOR
here is the order used to build the result vector(from text file)
FORMULA_NAME DNAwalker-PT-03track12BlockBoth-UpperBounds-00
FORMULA_NAME DNAwalker-PT-03track12BlockBoth-UpperBounds-01
FORMULA_NAME DNAwalker-PT-03track12BlockBoth-UpperBounds-02
FORMULA_NAME DNAwalker-PT-03track12BlockBoth-UpperBounds-03
FORMULA_NAME DNAwalker-PT-03track12BlockBoth-UpperBounds-04
FORMULA_NAME DNAwalker-PT-03track12BlockBoth-UpperBounds-05
FORMULA_NAME DNAwalker-PT-03track12BlockBoth-UpperBounds-06
FORMULA_NAME DNAwalker-PT-03track12BlockBoth-UpperBounds-07
FORMULA_NAME DNAwalker-PT-03track12BlockBoth-UpperBounds-08
FORMULA_NAME DNAwalker-PT-03track12BlockBoth-UpperBounds-09
FORMULA_NAME DNAwalker-PT-03track12BlockBoth-UpperBounds-10
FORMULA_NAME DNAwalker-PT-03track12BlockBoth-UpperBounds-11
FORMULA_NAME DNAwalker-PT-03track12BlockBoth-UpperBounds-12
FORMULA_NAME DNAwalker-PT-03track12BlockBoth-UpperBounds-13
FORMULA_NAME DNAwalker-PT-03track12BlockBoth-UpperBounds-14
FORMULA_NAME DNAwalker-PT-03track12BlockBoth-UpperBounds-15
=== Now, execution of the tool begins
BK_START 1678341337007
tapaal
Got BK_BIN_PATH=/home/mcc/BenchKit/bin/
---> tapaal --- TAPAAL
Setting MODEL_PATH=.
Setting VERIFYPN=/home/mcc/BenchKit/bin/verifypn
Got BK_TIME_CONFINEMENT=3600
Setting TEMPDIR=/home/mcc/BenchKit/bin/tmp
Got BK_MEMORY_CONFINEMENT=16384
Limiting to 16265216 kB
Total timeout: 3590
Time left: 3590
*****************************************
* TAPAAL CLASSIC verifying UpperBounds *
*****************************************
TEMPDIR=/home/mcc/BenchKit/bin/tmp
QF=/home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV
MF=/home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8
Time left: 3590
---------------------------------------------------
Step -1: Stripping Colors
---------------------------------------------------
Verifying stripped models (16 in total)
Time left: 3590
---------------------------------------------------
Step 0: Parallel Simplification
---------------------------------------------------
Doing parallel simplification (16 in total)
Total simplification timout is 718 -- reduction timeout is 299
timeout 3590 /home/mcc/BenchKit/bin/verifypn -n -q 718 -l 29 -d 299 -z 4 -s OverApprox --binary-query-io 2 --write-simplified /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --write-reduced /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 -x 1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16 ./model.pnml ./UpperBounds.xml
Time left: 3566
---------------------------------------------------
Step 1: Parallel processing
---------------------------------------------------
Doing parallel verification of individual queries (16 in total)
Each query is verified by 4 parallel strategies for 138 seconds
------------------- QUERY 1 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
Spent 0.001583 on verification
@@@0.00,5956@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --binary-query-io 1 -x 1 -n
FORMULA DNAwalker-PT-03track12BlockBoth-UpperBounds-00 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3566
------------------- QUERY 2 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
Spent 0.001303 on verification
@@@0.00,6052@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --binary-query-io 1 -x 2 -n
FORMULA DNAwalker-PT-03track12BlockBoth-UpperBounds-01 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3566
------------------- QUERY 3 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
Spent 0.001035 on verification
@@@0.00,5976@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --binary-query-io 1 -x 3 -n
FORMULA DNAwalker-PT-03track12BlockBoth-UpperBounds-02 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3565
------------------- QUERY 4 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
Spent 0.001277 on verification
@@@0.00,6036@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --binary-query-io 1 -x 4 -n
FORMULA DNAwalker-PT-03track12BlockBoth-UpperBounds-03 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3565
------------------- QUERY 5 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
Spent 0.001796 on verification
@@@0.00,6128@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --binary-query-io 1 -x 5 -n
FORMULA DNAwalker-PT-03track12BlockBoth-UpperBounds-04 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3565
------------------- QUERY 6 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
Spent 0.00172 on verification
@@@0.00,6284@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --binary-query-io 1 -x 6 -n
FORMULA DNAwalker-PT-03track12BlockBoth-UpperBounds-05 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3565
------------------- QUERY 7 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
Spent 0.001513 on verification
@@@0.00,5824@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --binary-query-io 1 -x 7 -n
FORMULA DNAwalker-PT-03track12BlockBoth-UpperBounds-06 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3565
------------------- QUERY 8 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
Spent 0.001616 on verification
@@@0.00,6256@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --binary-query-io 1 -x 8 -n
FORMULA DNAwalker-PT-03track12BlockBoth-UpperBounds-07 1 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3564
------------------- QUERY 9 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
Spent 0.000885 on verification
@@@0.00,6288@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --binary-query-io 1 -x 9 -n
FORMULA DNAwalker-PT-03track12BlockBoth-UpperBounds-08 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3564
------------------- QUERY 10 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
Spent 0.001048 on verification
@@@0.00,6188@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --binary-query-io 1 -x 10 -n
FORMULA DNAwalker-PT-03track12BlockBoth-UpperBounds-09 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3564
------------------- QUERY 11 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
Spent 0.00149 on verification
@@@0.00,6264@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --binary-query-io 1 -x 11 -n
FORMULA DNAwalker-PT-03track12BlockBoth-UpperBounds-10 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3564
------------------- QUERY 12 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
Spent 0.000982 on verification
@@@0.00,6172@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --binary-query-io 1 -x 12 -n
FORMULA DNAwalker-PT-03track12BlockBoth-UpperBounds-11 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION
Query index 0 was solved
Query is satisfied.
Spent 0.000848 on verification
@@@0.00,6176@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -p\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --binary-query-io 1 -x 12 -n
FORMULA DNAwalker-PT-03track12BlockBoth-UpperBounds-11 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3563
------------------- QUERY 13 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
Spent 0.001696 on verification
@@@0.00,6056@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --binary-query-io 1 -x 13 -n
FORMULA DNAwalker-PT-03track12BlockBoth-UpperBounds-12 1 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3563
------------------- QUERY 14 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
Spent 0.001518 on verification
@@@0.00,6168@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --binary-query-io 1 -x 14 -n
FORMULA DNAwalker-PT-03track12BlockBoth-UpperBounds-13 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3563
------------------- QUERY 15 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
Spent 0.001595 on verification
@@@0.00,6164@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --binary-query-io 1 -x 15 -n
FORMULA DNAwalker-PT-03track12BlockBoth-UpperBounds-14 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3563
------------------- QUERY 16 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "fr_FR.UTF8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
Spent 0.00177 on verification
@@@0.00,5996@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.UhWEhTBha8 /home/mcc/BenchKit/bin/tmp/tmp.5Cs219aFMV --binary-query-io 1 -x 16 -n
FORMULA DNAwalker-PT-03track12BlockBoth-UpperBounds-15 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3563
All queries are solved
Time left: 3563
terminated-with-cleanup
BK_STOP 1678341364977
--------------------
content from stderr:
CPN OverApproximation is only usable on colored models
Sequence of Actions to be Executed by the VM
This is useful if one wants to reexecute the tool in the VM from the submitted image disk.
set -x
# this is for BenchKit: configuration of major elements for the test
export BK_INPUT="DNAwalker-PT-03track12BlockBoth"
export BK_EXAMINATION="UpperBounds"
export BK_TOOL="tapaal"
export BK_RESULT_DIR="/tmp/BK_RESULTS/OUTPUTS"
export BK_TIME_CONFINEMENT="3600"
export BK_MEMORY_CONFINEMENT="16384"
export BK_BIN_PATH="/home/mcc/BenchKit/bin/"
# this is specific to your benchmark or test
export BIN_DIR="$HOME/BenchKit/bin"
# remove the execution directoty if it exists (to avoid increse of .vmdk images)
if [ -d execution ] ; then
rm -rf execution
fi
# this is for BenchKit: explicit launching of the test
echo "====================================================================="
echo " Generated by BenchKit 2-5348"
echo " Executing tool tapaal"
echo " Input is DNAwalker-PT-03track12BlockBoth, examination is UpperBounds"
echo " Time confinement is $BK_TIME_CONFINEMENT seconds"
echo " Memory confinement is 16384 MBytes"
echo " Number of cores is 4"
echo " Run identifier is r109-tall-167814487700893"
echo "====================================================================="
echo
echo "--------------------"
echo "preparation of the directory to be used:"
tar xzf /home/mcc/BenchKit/INPUTS/DNAwalker-PT-03track12BlockBoth.tgz
mv DNAwalker-PT-03track12BlockBoth execution
cd execution
if [ "UpperBounds" = "ReachabilityDeadlock" ] || [ "UpperBounds" = "UpperBounds" ] || [ "UpperBounds" = "QuasiLiveness" ] || [ "UpperBounds" = "StableMarking" ] || [ "UpperBounds" = "Liveness" ] || [ "UpperBounds" = "OneSafe" ] || [ "UpperBounds" = "StateSpace" ]; then
rm -f GenericPropertiesVerdict.xml
fi
pwd
ls -lh
echo
echo "--------------------"
echo "content from stdout:"
echo
echo "=== Data for post analysis generated by BenchKit (invocation template)"
echo
if [ "UpperBounds" = "UpperBounds" ] ; then
echo "The expected result is a vector of positive values"
echo NUM_VECTOR
elif [ "UpperBounds" != "StateSpace" ] ; then
echo "The expected result is a vector of booleans"
echo BOOL_VECTOR
else
echo "no data necessary for post analysis"
fi
echo
if [ -f "UpperBounds.txt" ] ; then
echo "here is the order used to build the result vector(from text file)"
for x in $(grep Property UpperBounds.txt | cut -d ' ' -f 2 | sort -u) ; do
echo "FORMULA_NAME $x"
done
elif [ -f "UpperBounds.xml" ] ; then # for cunf (txt files deleted;-)
echo echo "here is the order used to build the result vector(from xml file)"
for x in $(grep '
echo "FORMULA_NAME $x"
done
elif [ "UpperBounds" = "ReachabilityDeadlock" ] || [ "UpperBounds" = "QuasiLiveness" ] || [ "UpperBounds" = "StableMarking" ] || [ "UpperBounds" = "Liveness" ] || [ "UpperBounds" = "OneSafe" ] ; then
echo "FORMULA_NAME UpperBounds"
fi
echo
echo "=== Now, execution of the tool begins"
echo
echo -n "BK_START "
date -u +%s%3N
echo
timeout -s 9 $BK_TIME_CONFINEMENT bash -c "/home/mcc/BenchKit/BenchKit_head.sh 2> STDERR ; echo ; echo -n \"BK_STOP \" ; date -u +%s%3N"
if [ $? -eq 137 ] ; then
echo
echo "BK_TIME_CONFINEMENT_REACHED"
fi
echo
echo "--------------------"
echo "content from stderr:"
echo
cat STDERR ;

