About the Execution of ITS-Tools for DNAwalker-PT-16redondantChoiceR
| Execution Summary | |||||
| Max Memory Used (MB) |
Time wait (ms) | CPU Usage (ms) | I/O Wait (ms) | Computed Result | Execution Status |
| 642.787 | 88607.00 | 112252.00 | 393.60 | FFFFFTFFFTTFFFFF | normal |
Execution Chart
We display below the execution chart for this examination (boot time has been removed).
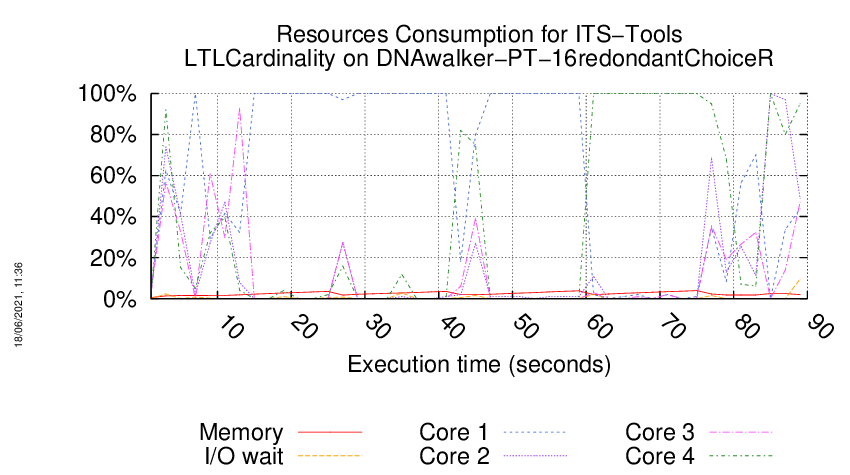
Trace from the execution
Formatting '/data/fkordon/mcc2021-input.r064-tall-162038397500652.qcow2', fmt=qcow2 size=4294967296 backing_file='/data/fkordon/mcc2021-input.qcow2' encryption=off cluster_size=65536 lazy_refcounts=off
Waiting for the VM to be ready (probing ssh)
............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................
=====================================================================
Generated by BenchKit 2-4028
Executing tool itstools
Input is DNAwalker-PT-16redondantChoiceR, examination is LTLCardinality
Time confinement is 3600 seconds
Memory confinement is 16384 MBytes
Number of cores is 4
Run identifier is r064-tall-162038397500652
=====================================================================
--------------------
preparation of the directory to be used:
/home/mcc/execution
total 596K
-rw-r--r-- 1 mcc users 13K May 5 16:58 CTLCardinality.txt
-rw-r--r-- 1 mcc users 129K May 5 16:58 CTLCardinality.xml
-rw-r--r-- 1 mcc users 12K May 5 16:58 CTLFireability.txt
-rw-r--r-- 1 mcc users 91K May 5 16:58 CTLFireability.xml
-rw-r--r-- 1 mcc users 4.2K May 6 14:48 GenericPropertiesDefinition.xml
-rw-r--r-- 1 mcc users 6.0K May 6 14:48 GenericPropertiesVerdict.xml
-rw-r--r-- 1 mcc users 3.4K Mar 28 16:00 LTLCardinality.txt
-rw-r--r-- 1 mcc users 24K Mar 28 16:00 LTLCardinality.xml
-rw-r--r-- 1 mcc users 2.3K Mar 28 16:00 LTLFireability.txt
-rw-r--r-- 1 mcc users 18K Mar 28 16:00 LTLFireability.xml
-rw-r--r-- 1 mcc users 3.7K Mar 23 10:03 ReachabilityCardinality.txt
-rw-r--r-- 1 mcc users 18K Mar 23 10:03 ReachabilityCardinality.xml
-rw-r--r-- 1 mcc users 2.9K Mar 22 17:37 ReachabilityFireability.txt
-rw-r--r-- 1 mcc users 15K Mar 22 17:37 ReachabilityFireability.xml
-rw-r--r-- 1 mcc users 1.8K Mar 22 09:10 UpperBounds.txt
-rw-r--r-- 1 mcc users 3.9K Mar 22 09:10 UpperBounds.xml
-rw-r--r-- 1 mcc users 6 May 5 16:51 equiv_col
-rw-r--r-- 1 mcc users 19 May 5 16:51 instance
-rw-r--r-- 1 mcc users 6 May 5 16:51 iscolored
-rw-r--r-- 1 mcc users 211K May 5 16:51 model.pnml
--------------------
content from stdout:
=== Data for post analysis generated by BenchKit (invocation template)
The expected result is a vector of booleans
BOOL_VECTOR
here is the order used to build the result vector(from text file)
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-00
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-01
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-02
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-03
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-04
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-05
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-06
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-07
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-08
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-09
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-10
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-11
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-12
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-13
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-14
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-15
=== Now, execution of the tool begins
BK_START 1620548808117
Running Version 0
[2021-05-09 08:26:49] [INFO ] Running its-tools with arguments : [-pnfolder, /home/mcc/execution, -examination, LTLCardinality, -spotpath, /home/mcc/BenchKit/bin//..//ltlfilt, -z3path, /home/mcc/BenchKit/bin//..//z3/bin/z3, -yices2path, /home/mcc/BenchKit/bin//..//yices/bin/yices, -its, -ltsmin, -greatspnpath, /home/mcc/BenchKit/bin//..//greatspn/, -order, META, -manyOrder, -smt, -timeout, 3600]
[2021-05-09 08:26:49] [INFO ] Parsing pnml file : /home/mcc/execution/model.pnml
[2021-05-09 08:26:49] [INFO ] Load time of PNML (sax parser for PT used): 70 ms
[2021-05-09 08:26:49] [INFO ] Transformed 43 places.
[2021-05-09 08:26:49] [INFO ] Transformed 490 transitions.
[2021-05-09 08:26:49] [INFO ] Parsed PT model containing 43 places and 490 transitions in 110 ms.
Parsed 16 properties from file /home/mcc/execution/LTLCardinality.xml in 9 ms.
Working with output stream class java.io.PrintStream
[2021-05-09 08:26:49] [INFO ] Initial state test concluded for 1 properties.
FORMULA DNAwalker-PT-16redondantChoiceR-05 TRUE TECHNIQUES TOPOLOGICAL INITIAL_STATE
FORMULA DNAwalker-PT-16redondantChoiceR-08 FALSE TECHNIQUES TOPOLOGICAL INITIAL_STATE
FORMULA DNAwalker-PT-16redondantChoiceR-10 TRUE TECHNIQUES TOPOLOGICAL INITIAL_STATE
Support contains 28 out of 43 places. Attempting structural reductions.
Starting structural reductions, iteration 0 : 43/43 places, 490/490 transitions.
Applied a total of 0 rules in 16 ms. Remains 43 /43 variables (removed 0) and now considering 490/490 (removed 0) transitions.
[2021-05-09 08:26:49] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:26:49] [INFO ] Computed 0 place invariants in 16 ms
[2021-05-09 08:26:50] [INFO ] Dead Transitions using invariants and state equation in 291 ms returned []
[2021-05-09 08:26:50] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:26:50] [INFO ] Computed 0 place invariants in 4 ms
[2021-05-09 08:26:50] [INFO ] Implicit Places using invariants in 60 ms returned []
[2021-05-09 08:26:50] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:26:50] [INFO ] Computed 0 place invariants in 9 ms
[2021-05-09 08:26:50] [INFO ] State equation strengthened by 161 read => feed constraints.
[2021-05-09 08:26:50] [INFO ] Implicit Places using invariants and state equation in 284 ms returned []
Implicit Place search using SMT with State Equation took 346 ms to find 0 implicit places.
[2021-05-09 08:26:50] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:26:50] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:26:50] [INFO ] Dead Transitions using invariants and state equation in 160 ms returned []
Finished structural reductions, in 1 iterations. Remains : 43/43 places, 490/490 transitions.
[2021-05-09 08:26:50] [INFO ] Initial state reduction rules for LTL removed 2 formulas.
[2021-05-09 08:26:50] [INFO ] Flatten gal took : 60 ms
FORMULA DNAwalker-PT-16redondantChoiceR-12 FALSE TECHNIQUES TOPOLOGICAL INITIAL_STATE
FORMULA DNAwalker-PT-16redondantChoiceR-09 TRUE TECHNIQUES TOPOLOGICAL INITIAL_STATE
[2021-05-09 08:26:50] [INFO ] Flatten gal took : 27 ms
[2021-05-09 08:26:50] [INFO ] Input system was already deterministic with 490 transitions.
Incomplete random walk after 100000 steps, including 3826 resets, run finished after 597 ms. (steps per millisecond=167 ) properties (out of 23) seen :21
Running SMT prover for 2 properties.
[2021-05-09 08:26:51] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:26:51] [INFO ] Computed 0 place invariants in 2 ms
[2021-05-09 08:26:51] [INFO ] [Real]Adding state equation constraints to refine reachable states.
[2021-05-09 08:26:51] [INFO ] [Real]Absence check using state equation in 59 ms returned unsat
[2021-05-09 08:26:51] [INFO ] [Real]Adding state equation constraints to refine reachable states.
[2021-05-09 08:26:51] [INFO ] [Real]Absence check using state equation in 63 ms returned sat
[2021-05-09 08:26:51] [INFO ] Solution in real domain found non-integer solution.
[2021-05-09 08:26:51] [INFO ] [Nat]Adding state equation constraints to refine reachable states.
[2021-05-09 08:26:55] [INFO ] [Nat]Absence check using state equation in 3252 ms returned unsat
Successfully simplified 2 atomic propositions for a total of 11 simplifications.
[2021-05-09 08:26:55] [INFO ] Initial state test concluded for 1 properties.
FORMULA DNAwalker-PT-16redondantChoiceR-03 FALSE TECHNIQUES TOPOLOGICAL INITIAL_STATE
Running Spot : CommandLine [args=[/home/mcc/BenchKit/bin//..//ltl2tgba, --hoaf=tv, -f, !(F(G(p0)))], workingDir=/home/mcc/execution]
Support contains 2 out of 43 places. Attempting structural reductions.
Starting structural reductions, iteration 0 : 43/43 places, 490/490 transitions.
Applied a total of 0 rules in 26 ms. Remains 43 /43 variables (removed 0) and now considering 490/490 (removed 0) transitions.
[2021-05-09 08:26:55] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:26:55] [INFO ] Computed 0 place invariants in 10 ms
[2021-05-09 08:26:55] [INFO ] Dead Transitions using invariants and state equation in 210 ms returned []
[2021-05-09 08:26:55] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:26:55] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:26:55] [INFO ] Implicit Places using invariants in 52 ms returned []
[2021-05-09 08:26:55] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:26:55] [INFO ] Computed 0 place invariants in 4 ms
[2021-05-09 08:26:55] [INFO ] State equation strengthened by 161 read => feed constraints.
[2021-05-09 08:26:55] [INFO ] Implicit Places using invariants and state equation in 534 ms returned []
Implicit Place search using SMT with State Equation took 588 ms to find 0 implicit places.
[2021-05-09 08:26:55] [INFO ] Redundant transitions in 32 ms returned []
[2021-05-09 08:26:55] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:26:55] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:26:56] [INFO ] Dead Transitions using invariants and state equation in 147 ms returned []
Finished structural reductions, in 1 iterations. Remains : 43/43 places, 490/490 transitions.
Stuttering acceptance computed with spot in 134 ms :[(NOT p0)]
Running random walk in product with property : DNAwalker-PT-16redondantChoiceR-00 automaton TGBA [mat=[[{ cond=(NOT p0), acceptance={0} source=0 dest: 0}, { cond=p0, acceptance={} source=0 dest: 0}]], initial=0, aps=[p0:(GT s18 s10)], nbAcceptance=1, properties=[trans-labels, explicit-labels, trans-acc, complete, deterministic, no-univ-branch, unambiguous, semi-deterministic, stutter-invariant], stateDesc=[null][true]]
Product exploration explored 100000 steps with 0 reset in 205 ms.
Stuttering criterion allowed to conclude after 28 steps with 0 reset in 1 ms.
FORMULA DNAwalker-PT-16redondantChoiceR-00 FALSE TECHNIQUES STUTTER_TEST
Treatment of property DNAwalker-PT-16redondantChoiceR-00 finished in 1409 ms.
Running Spot : CommandLine [args=[/home/mcc/BenchKit/bin//..//ltl2tgba, --hoaf=tv, -f, !((F(p0) U (G(F(p1))||(p2&&F(p1)))))], workingDir=/home/mcc/execution]
Support contains 4 out of 43 places. Attempting structural reductions.
Starting structural reductions, iteration 0 : 43/43 places, 490/490 transitions.
Applied a total of 0 rules in 16 ms. Remains 43 /43 variables (removed 0) and now considering 490/490 (removed 0) transitions.
[2021-05-09 08:26:56] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:26:56] [INFO ] Computed 0 place invariants in 2 ms
[2021-05-09 08:26:56] [INFO ] Dead Transitions using invariants and state equation in 162 ms returned []
[2021-05-09 08:26:56] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:26:56] [INFO ] Computed 0 place invariants in 2 ms
[2021-05-09 08:26:56] [INFO ] Implicit Places using invariants in 37 ms returned []
[2021-05-09 08:26:56] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:26:56] [INFO ] Computed 0 place invariants in 2 ms
[2021-05-09 08:26:56] [INFO ] State equation strengthened by 161 read => feed constraints.
[2021-05-09 08:26:57] [INFO ] Implicit Places using invariants and state equation in 892 ms returned []
Implicit Place search using SMT with State Equation took 931 ms to find 0 implicit places.
[2021-05-09 08:26:57] [INFO ] Redundant transitions in 40 ms returned []
[2021-05-09 08:26:57] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:26:57] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:26:57] [INFO ] Dead Transitions using invariants and state equation in 150 ms returned []
Finished structural reductions, in 1 iterations. Remains : 43/43 places, 490/490 transitions.
Stuttering acceptance computed with spot in 127 ms :[(NOT p1), (NOT p1), (AND (NOT p1) (NOT p0)), (AND (NOT p1) (NOT p0))]
Running random walk in product with property : DNAwalker-PT-16redondantChoiceR-01 automaton TGBA [mat=[[{ cond=(NOT p2), acceptance={} source=0 dest: 0}, { cond=(NOT p1), acceptance={} source=0 dest: 1}, { cond=(AND (NOT p2) (NOT p0)), acceptance={} source=0 dest: 3}], [{ cond=(NOT p1), acceptance={0} source=1 dest: 1}], [{ cond=(AND (NOT p1) (NOT p0)), acceptance={0} source=2 dest: 2}], [{ cond=(AND (NOT p1) (NOT p0)), acceptance={} source=3 dest: 2}, { cond=(NOT p0), acceptance={} source=3 dest: 3}]], initial=0, aps=[p2:(GT s18 s0), p1:(OR (AND (OR (GT s18 s0) (LEQ 1 s20) (GT 1 s29)) (GT 1 s29)) (LEQ s18 s0)), p0:(OR (LEQ s18 s0) (GT 1 s29))], nbAcceptance=1, properties=[trans-labels, explicit-labels, trans-acc, no-univ-branch, stutter-invariant], stateDesc=[null, null, null, null][true, true, true, true]]
Product exploration explored 100000 steps with 2 reset in 129 ms.
Product exploration explored 100000 steps with 0 reset in 149 ms.
Knowledge obtained : [(NOT p2), p1, p0]
Stuttering acceptance computed with spot in 98 ms :[(NOT p1), (NOT p1), (AND (NOT p1) (NOT p0)), (AND (NOT p1) (NOT p0))]
Product exploration explored 100000 steps with 0 reset in 97 ms.
Product exploration explored 100000 steps with 0 reset in 187 ms.
[2021-05-09 08:26:58] [INFO ] Flatten gal took : 22 ms
[2021-05-09 08:26:58] [INFO ] Flatten gal took : 21 ms
[2021-05-09 08:26:58] [INFO ] Time to serialize gal into /tmp/LTL11682139907225725024.gal : 5 ms
[2021-05-09 08:26:58] [INFO ] Time to serialize properties into /tmp/LTL5312038988567107298.ltl : 1 ms
Invoking ITS tools like this :CommandLine [args=[/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.itstools.binaries_1.0.0.202104292328/bin/its-ltl-linux64, --gc-threshold, 2000000, -i, /tmp/LTL11682139907225725024.gal, -t, CGAL, -LTL, /tmp/LTL5312038988567107298.ltl, -c, -stutter-deadlock, --gen-order, FOLLOW], workingDir=/home/mcc/execution]
its-ltl command run as :
/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.itstools.binaries_1.0.0.202104292328/bin/its-ltl-linux64 --gc-threshold 2000000 -i /tmp/LTL11682139907225725024.gal -t CGAL -LTL /tmp/LTL5312038988567107298.ltl -c -stutter-deadlock --gen-order FOLLOW
Read 1 LTL properties
Checking formula 0 : !(((F("((A15<=A1)||(A24<1))"))U((G(F("(((((A15>A1)||(A16>=1))||(A24<1))&&(A24<1))||(A15<=A1))")))||(("(A15>A1)")&&(F("(((((A15>A1)||(A16>=1))||(A24<1))&&(A24<1))||(A15<=A1))"))))))
Formula 0 simplified : !(F"((A15<=A1)||(A24<1))" U (GF"(((((A15>A1)||(A16>=1))||(A24<1))&&(A24<1))||(A15<=A1))" | ("(A15>A1)" & F"(((((A15>A1)||(A16>=1))||(A24<1))&&(A24<1))||(A15<=A1))")))
Detected timeout of ITS tools.
[2021-05-09 08:27:13] [INFO ] Flatten gal took : 46 ms
[2021-05-09 08:27:13] [INFO ] Applying decomposition
[2021-05-09 08:27:13] [INFO ] Flatten gal took : 19 ms
Converted graph to binary with : CommandLine [args=[/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.louvain.binaries_1.0.0.202104292328/bin/convert-linux64, -i, /tmp/graph4237358786691456883.txt, -o, /tmp/graph4237358786691456883.bin, -w, /tmp/graph4237358786691456883.weights], workingDir=null]
Built communities with : CommandLine [args=[/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.louvain.binaries_1.0.0.202104292328/bin/louvain-linux64, /tmp/graph4237358786691456883.bin, -l, -1, -v, -w, /tmp/graph4237358786691456883.weights, -q, 0, -e, 0.001], workingDir=null]
[2021-05-09 08:27:14] [INFO ] Decomposing Gal with order
[2021-05-09 08:27:14] [INFO ] Rewriting arrays to variables to allow decomposition.
[2021-05-09 08:27:14] [INFO ] Removed a total of 872 redundant transitions.
[2021-05-09 08:27:14] [INFO ] Flatten gal took : 181 ms
[2021-05-09 08:27:14] [INFO ] Fuse similar labels procedure discarded/fused a total of 0 labels/synchronizations in 5 ms.
[2021-05-09 08:27:14] [INFO ] Time to serialize gal into /tmp/LTL16185481032411871162.gal : 3 ms
[2021-05-09 08:27:14] [INFO ] Time to serialize properties into /tmp/LTL18331227636983109252.ltl : 0 ms
Invoking ITS tools like this :CommandLine [args=[/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.itstools.binaries_1.0.0.202104292328/bin/its-ltl-linux64, --gc-threshold, 2000000, -i, /tmp/LTL16185481032411871162.gal, -t, CGAL, -LTL, /tmp/LTL18331227636983109252.ltl, -c, -stutter-deadlock, --gen-order, FOLLOW], workingDir=/home/mcc/execution]
its-ltl command run as :
/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.itstools.binaries_1.0.0.202104292328/bin/its-ltl-linux64 --gc-threshold 2000000 -i /tmp/LTL16185481032411871162.gal -t CGAL -LTL /tmp/LTL18331227636983109252.ltl -c -stutter-deadlock --gen-order FOLLOW
Read 1 LTL properties
Checking formula 0 : !(((F("((gu13.A15<=gu13.A1)||(gu22.A24<1))"))U((G(F("(((((gu13.A15>gu13.A1)||(gu14.A16>=1))||(gu22.A24<1))&&(gu22.A24<1))||(gu13.A15<=gu13.A1))")))||(("(gu13.A15>gu13.A1)")&&(F("(((((gu13.A15>gu13.A1)||(gu14.A16>=1))||(gu22.A24<1))&&(gu22.A24<1))||(gu13.A15<=gu13.A1))"))))))
Formula 0 simplified : !(F"((gu13.A15<=gu13.A1)||(gu22.A24<1))" U (GF"(((((gu13.A15>gu13.A1)||(gu14.A16>=1))||(gu22.A24<1))&&(gu22.A24<1))||(gu13.A15<=gu13.A1))" | ("(gu13.A15>gu13.A1)" & F"(((((gu13.A15>gu13.A1)||(gu14.A16>=1))||(gu22.A24<1))&&(gu22.A24<1))||(gu13.A15<=gu13.A1))")))
Detected timeout of ITS tools.
Built C files in :
/tmp/ltsmin9813753424145887906
[2021-05-09 08:27:29] [INFO ] Built C files in 26ms conformant to PINS (ltsmin variant)in folder :/tmp/ltsmin9813753424145887906
Running compilation step : CommandLine [args=[gcc, -c, -I/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.ltsmin.binaries_1.0.0.202104292328/bin/include/, -I., -std=c99, -fPIC, -O2, model.c], workingDir=/tmp/ltsmin9813753424145887906]
WARNING : LTS min runner thread failed on error :java.lang.RuntimeException: Compilation or link of executable timed out.java.util.concurrent.TimeoutException: Subprocess running CommandLine [args=[gcc, -c, -I/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.ltsmin.binaries_1.0.0.202104292328/bin/include/, -I., -std=c99, -fPIC, -O2, model.c], workingDir=/tmp/ltsmin9813753424145887906] killed by timeout after 1 SECONDS
java.lang.RuntimeException: Compilation or link of executable timed out.java.util.concurrent.TimeoutException: Subprocess running CommandLine [args=[gcc, -c, -I/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.ltsmin.binaries_1.0.0.202104292328/bin/include/, -I., -std=c99, -fPIC, -O2, model.c], workingDir=/tmp/ltsmin9813753424145887906] killed by timeout after 1 SECONDS
at fr.lip6.move.gal.application.LTSminRunner$1.run(LTSminRunner.java:79)
at java.base/java.lang.Thread.run(Thread.java:834)
Treatment of property DNAwalker-PT-16redondantChoiceR-01 finished in 33880 ms.
Running Spot : CommandLine [args=[/home/mcc/BenchKit/bin//..//ltl2tgba, --hoaf=tv, -f, !((p0 U G(p1)))], workingDir=/home/mcc/execution]
Support contains 4 out of 43 places. Attempting structural reductions.
Starting structural reductions, iteration 0 : 43/43 places, 490/490 transitions.
Applied a total of 0 rules in 14 ms. Remains 43 /43 variables (removed 0) and now considering 490/490 (removed 0) transitions.
[2021-05-09 08:27:30] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:27:30] [INFO ] Computed 0 place invariants in 2 ms
[2021-05-09 08:27:30] [INFO ] Dead Transitions using invariants and state equation in 159 ms returned []
[2021-05-09 08:27:30] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:27:30] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:27:30] [INFO ] Implicit Places using invariants in 36 ms returned []
[2021-05-09 08:27:30] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:27:30] [INFO ] Computed 0 place invariants in 8 ms
[2021-05-09 08:27:30] [INFO ] State equation strengthened by 161 read => feed constraints.
[2021-05-09 08:27:31] [INFO ] Implicit Places using invariants and state equation in 630 ms returned []
Implicit Place search using SMT with State Equation took 667 ms to find 0 implicit places.
[2021-05-09 08:27:31] [INFO ] Redundant transitions in 3 ms returned []
[2021-05-09 08:27:31] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:27:31] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:27:31] [INFO ] Dead Transitions using invariants and state equation in 173 ms returned []
Finished structural reductions, in 1 iterations. Remains : 43/43 places, 490/490 transitions.
Stuttering acceptance computed with spot in 60 ms :[(NOT p1), true, (NOT p1)]
Running random walk in product with property : DNAwalker-PT-16redondantChoiceR-02 automaton TGBA [mat=[[{ cond=(AND (NOT p1) p0), acceptance={0} source=0 dest: 0}, { cond=(AND p1 p0), acceptance={} source=0 dest: 0}, { cond=(AND (NOT p1) (NOT p0)), acceptance={} source=0 dest: 1}, { cond=(AND p1 (NOT p0)), acceptance={} source=0 dest: 2}], [{ cond=true, acceptance={0} source=1 dest: 1}], [{ cond=(NOT p1), acceptance={} source=2 dest: 1}, { cond=p1, acceptance={} source=2 dest: 2}]], initial=0, aps=[p1:(LEQ s40 s27), p0:(AND (LEQ s40 s27) (GT s3 s10))], nbAcceptance=1, properties=[trans-labels, explicit-labels, trans-acc, complete, deterministic, no-univ-branch, unambiguous, semi-deterministic, stutter-invariant], stateDesc=[null, null, null][true, true, true]]
Product exploration explored 100000 steps with 0 reset in 87 ms.
Product exploration explored 100000 steps with 0 reset in 81 ms.
Knowledge obtained : [p1, (NOT p0)]
Stuttering acceptance computed with spot in 65 ms :[(NOT p1), true, (NOT p1)]
Product exploration explored 100000 steps with 1 reset in 71 ms.
Product exploration explored 100000 steps with 0 reset in 89 ms.
[2021-05-09 08:27:31] [INFO ] Flatten gal took : 19 ms
[2021-05-09 08:27:31] [INFO ] Flatten gal took : 18 ms
[2021-05-09 08:27:31] [INFO ] Time to serialize gal into /tmp/LTL6963420256180123773.gal : 2 ms
[2021-05-09 08:27:31] [INFO ] Time to serialize properties into /tmp/LTL6349862825386626606.ltl : 0 ms
Invoking ITS tools like this :CommandLine [args=[/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.itstools.binaries_1.0.0.202104292328/bin/its-ltl-linux64, --gc-threshold, 2000000, -i, /tmp/LTL6963420256180123773.gal, -t, CGAL, -LTL, /tmp/LTL6349862825386626606.ltl, -c, -stutter-deadlock, --gen-order, FOLLOW], workingDir=/home/mcc/execution]
its-ltl command run as :
/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.itstools.binaries_1.0.0.202104292328/bin/its-ltl-linux64 --gc-threshold 2000000 -i /tmp/LTL6963420256180123773.gal -t CGAL -LTL /tmp/LTL6349862825386626606.ltl -c -stutter-deadlock --gen-order FOLLOW
Read 1 LTL properties
Checking formula 0 : !((("((B31<=A22)&&(A4>A9))")U(G("(B31<=A22)"))))
Formula 0 simplified : !("((B31<=A22)&&(A4>A9))" U G"(B31<=A22)")
Detected timeout of ITS tools.
[2021-05-09 08:27:47] [INFO ] Flatten gal took : 16 ms
[2021-05-09 08:27:47] [INFO ] Applying decomposition
[2021-05-09 08:27:47] [INFO ] Flatten gal took : 15 ms
Converted graph to binary with : CommandLine [args=[/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.louvain.binaries_1.0.0.202104292328/bin/convert-linux64, -i, /tmp/graph11113675083606912473.txt, -o, /tmp/graph11113675083606912473.bin, -w, /tmp/graph11113675083606912473.weights], workingDir=null]
Built communities with : CommandLine [args=[/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.louvain.binaries_1.0.0.202104292328/bin/louvain-linux64, /tmp/graph11113675083606912473.bin, -l, -1, -v, -w, /tmp/graph11113675083606912473.weights, -q, 0, -e, 0.001], workingDir=null]
[2021-05-09 08:27:47] [INFO ] Decomposing Gal with order
[2021-05-09 08:27:47] [INFO ] Rewriting arrays to variables to allow decomposition.
[2021-05-09 08:27:47] [INFO ] Removed a total of 868 redundant transitions.
[2021-05-09 08:27:47] [INFO ] Flatten gal took : 37 ms
[2021-05-09 08:27:47] [INFO ] Fuse similar labels procedure discarded/fused a total of 0 labels/synchronizations in 3 ms.
[2021-05-09 08:27:47] [INFO ] Time to serialize gal into /tmp/LTL3312767530590419331.gal : 3 ms
[2021-05-09 08:27:47] [INFO ] Time to serialize properties into /tmp/LTL15108707534061403233.ltl : 0 ms
Invoking ITS tools like this :CommandLine [args=[/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.itstools.binaries_1.0.0.202104292328/bin/its-ltl-linux64, --gc-threshold, 2000000, -i, /tmp/LTL3312767530590419331.gal, -t, CGAL, -LTL, /tmp/LTL15108707534061403233.ltl, -c, -stutter-deadlock, --gen-order, FOLLOW], workingDir=/home/mcc/execution]
its-ltl command run as :
/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.itstools.binaries_1.0.0.202104292328/bin/its-ltl-linux64 --gc-threshold 2000000 -i /tmp/LTL3312767530590419331.gal -t CGAL -LTL /tmp/LTL15108707534061403233.ltl -c -stutter-deadlock --gen-order FOLLOW
Read 1 LTL properties
Checking formula 0 : !((("((gu20.B31<=gu20.A22)&&(gu7.A4>gu7.A9))")U(G("(gu20.B31<=gu20.A22)"))))
Formula 0 simplified : !("((gu20.B31<=gu20.A22)&&(gu7.A4>gu7.A9))" U G"(gu20.B31<=gu20.A22)")
Detected timeout of ITS tools.
Built C files in :
/tmp/ltsmin17989254189611288703
[2021-05-09 08:28:02] [INFO ] Built C files in 5ms conformant to PINS (ltsmin variant)in folder :/tmp/ltsmin17989254189611288703
Running compilation step : CommandLine [args=[gcc, -c, -I/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.ltsmin.binaries_1.0.0.202104292328/bin/include/, -I., -std=c99, -fPIC, -O2, model.c], workingDir=/tmp/ltsmin17989254189611288703]
WARNING : LTS min runner thread failed on error :java.lang.RuntimeException: Compilation or link of executable timed out.java.util.concurrent.TimeoutException: Subprocess running CommandLine [args=[gcc, -c, -I/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.ltsmin.binaries_1.0.0.202104292328/bin/include/, -I., -std=c99, -fPIC, -O2, model.c], workingDir=/tmp/ltsmin17989254189611288703] killed by timeout after 1 SECONDS
java.lang.RuntimeException: Compilation or link of executable timed out.java.util.concurrent.TimeoutException: Subprocess running CommandLine [args=[gcc, -c, -I/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.ltsmin.binaries_1.0.0.202104292328/bin/include/, -I., -std=c99, -fPIC, -O2, model.c], workingDir=/tmp/ltsmin17989254189611288703] killed by timeout after 1 SECONDS
at fr.lip6.move.gal.application.LTSminRunner$1.run(LTSminRunner.java:79)
at java.base/java.lang.Thread.run(Thread.java:834)
Treatment of property DNAwalker-PT-16redondantChoiceR-02 finished in 32859 ms.
Running Spot : CommandLine [args=[/home/mcc/BenchKit/bin//..//ltl2tgba, --hoaf=tv, -f, !(X((p0 U G((p1&&X((p0 U p1)))))))], workingDir=/home/mcc/execution]
Support contains 3 out of 43 places. Attempting structural reductions.
Starting structural reductions, iteration 0 : 43/43 places, 490/490 transitions.
Applied a total of 0 rules in 2 ms. Remains 43 /43 variables (removed 0) and now considering 490/490 (removed 0) transitions.
[2021-05-09 08:28:03] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:03] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:03] [INFO ] Dead Transitions using invariants and state equation in 187 ms returned []
[2021-05-09 08:28:03] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:03] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:03] [INFO ] Implicit Places using invariants in 40 ms returned []
[2021-05-09 08:28:03] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:03] [INFO ] Computed 0 place invariants in 2 ms
[2021-05-09 08:28:03] [INFO ] State equation strengthened by 161 read => feed constraints.
[2021-05-09 08:28:03] [INFO ] Implicit Places using invariants and state equation in 500 ms returned []
Implicit Place search using SMT with State Equation took 542 ms to find 0 implicit places.
[2021-05-09 08:28:03] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:03] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:04] [INFO ] Dead Transitions using invariants and state equation in 136 ms returned []
Finished structural reductions, in 1 iterations. Remains : 43/43 places, 490/490 transitions.
Stuttering acceptance computed with spot in 115 ms :[(NOT p1), (NOT p1), true, (NOT p1), (AND (NOT p1) p0)]
Running random walk in product with property : DNAwalker-PT-16redondantChoiceR-04 automaton TGBA [mat=[[{ cond=true, acceptance={} source=0 dest: 1}], [{ cond=(AND (NOT p1) p0), acceptance={0} source=1 dest: 1}, { cond=(AND p1 p0), acceptance={} source=1 dest: 1}, { cond=(AND (NOT p1) (NOT p0)), acceptance={} source=1 dest: 2}, { cond=p1, acceptance={} source=1 dest: 3}, { cond=(AND p1 (NOT p0)), acceptance={} source=1 dest: 4}], [{ cond=true, acceptance={0} source=2 dest: 2}], [{ cond=(AND (NOT p1) (NOT p0)), acceptance={} source=3 dest: 2}, { cond=(AND (NOT p1) p0), acceptance={0} source=3 dest: 3}], [{ cond=(AND (NOT p1) p0), acceptance={} source=4 dest: 2}, { cond=p1, acceptance={} source=4 dest: 3}, { cond=p1, acceptance={} source=4 dest: 4}]], initial=0, aps=[p1:(LEQ s18 s17), p0:(LEQ 1 s27)], nbAcceptance=1, properties=[trans-labels, explicit-labels, trans-acc, no-univ-branch], stateDesc=[null, null, null, null, null][false, false, false, false, false]]
Entered a terminal (fully accepting) state of product in 565 steps with 140 reset in 4 ms.
FORMULA DNAwalker-PT-16redondantChoiceR-04 FALSE TECHNIQUES STUTTER_TEST
Treatment of property DNAwalker-PT-16redondantChoiceR-04 finished in 1014 ms.
Running Spot : CommandLine [args=[/home/mcc/BenchKit/bin//..//ltl2tgba, --hoaf=tv, -f, !(X(F(p0)))], workingDir=/home/mcc/execution]
Support contains 2 out of 43 places. Attempting structural reductions.
Starting structural reductions, iteration 0 : 43/43 places, 490/490 transitions.
Applied a total of 0 rules in 2 ms. Remains 43 /43 variables (removed 0) and now considering 490/490 (removed 0) transitions.
[2021-05-09 08:28:04] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:04] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:04] [INFO ] Dead Transitions using invariants and state equation in 158 ms returned []
[2021-05-09 08:28:04] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:04] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:04] [INFO ] Implicit Places using invariants in 34 ms returned []
[2021-05-09 08:28:04] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:04] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:04] [INFO ] State equation strengthened by 161 read => feed constraints.
[2021-05-09 08:28:04] [INFO ] Implicit Places using invariants and state equation in 532 ms returned []
Implicit Place search using SMT with State Equation took 569 ms to find 0 implicit places.
[2021-05-09 08:28:04] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:04] [INFO ] Computed 0 place invariants in 2 ms
[2021-05-09 08:28:05] [INFO ] Dead Transitions using invariants and state equation in 133 ms returned []
Finished structural reductions, in 1 iterations. Remains : 43/43 places, 490/490 transitions.
Stuttering acceptance computed with spot in 51 ms :[(NOT p0), (NOT p0)]
Running random walk in product with property : DNAwalker-PT-16redondantChoiceR-06 automaton TGBA [mat=[[{ cond=true, acceptance={} source=0 dest: 1}], [{ cond=(NOT p0), acceptance={0} source=1 dest: 1}]], initial=0, aps=[p0:(GT s41 s12)], nbAcceptance=1, properties=[trans-labels, explicit-labels, trans-acc, deterministic, no-univ-branch, unambiguous, semi-deterministic, very-weak, weak, inherently-weak], stateDesc=[null, null][false, false]]
Stuttering criterion allowed to conclude after 104 steps with 4 reset in 1 ms.
FORMULA DNAwalker-PT-16redondantChoiceR-06 FALSE TECHNIQUES STUTTER_TEST
Treatment of property DNAwalker-PT-16redondantChoiceR-06 finished in 928 ms.
Running Spot : CommandLine [args=[/home/mcc/BenchKit/bin//..//ltl2tgba, --hoaf=tv, -f, !(X((G(F(p0))&&(G(p1)||(p0 U p2)))))], workingDir=/home/mcc/execution]
Support contains 4 out of 43 places. Attempting structural reductions.
Starting structural reductions, iteration 0 : 43/43 places, 490/490 transitions.
Applied a total of 0 rules in 3 ms. Remains 43 /43 variables (removed 0) and now considering 490/490 (removed 0) transitions.
[2021-05-09 08:28:05] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:05] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:05] [INFO ] Dead Transitions using invariants and state equation in 136 ms returned []
[2021-05-09 08:28:05] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:05] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:05] [INFO ] Implicit Places using invariants in 35 ms returned []
[2021-05-09 08:28:05] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:05] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:05] [INFO ] State equation strengthened by 161 read => feed constraints.
[2021-05-09 08:28:05] [INFO ] Implicit Places using invariants and state equation in 584 ms returned []
Implicit Place search using SMT with State Equation took 624 ms to find 0 implicit places.
[2021-05-09 08:28:05] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:05] [INFO ] Computed 0 place invariants in 2 ms
[2021-05-09 08:28:06] [INFO ] Dead Transitions using invariants and state equation in 138 ms returned []
Finished structural reductions, in 1 iterations. Remains : 43/43 places, 490/490 transitions.
Stuttering acceptance computed with spot in 182 ms :[(OR (NOT p0) (AND (NOT p1) (NOT p2))), (OR (NOT p0) (AND (NOT p1) (NOT p2))), true, (NOT p0), (NOT p0), (NOT p1), (NOT p2), (AND (NOT p1) (NOT p2))]
Running random walk in product with property : DNAwalker-PT-16redondantChoiceR-07 automaton TGBA [mat=[[{ cond=true, acceptance={} source=0 dest: 1}], [{ cond=(AND (NOT p0) (NOT p1) (NOT p2)), acceptance={} source=1 dest: 2}, { cond=(OR (AND (NOT p0) p1) (AND (NOT p0) p2)), acceptance={} source=1 dest: 3}, { cond=(OR p0 p1 p2), acceptance={} source=1 dest: 4}, { cond=(AND (NOT p0) p1 (NOT p2)), acceptance={} source=1 dest: 5}, { cond=(AND p0 (NOT p1) (NOT p2)), acceptance={} source=1 dest: 6}, { cond=(AND p0 p1 (NOT p2)), acceptance={} source=1 dest: 7}], [{ cond=true, acceptance={0} source=2 dest: 2}], [{ cond=(NOT p0), acceptance={0} source=3 dest: 3}], [{ cond=(NOT p0), acceptance={} source=4 dest: 3}, { cond=true, acceptance={} source=4 dest: 4}], [{ cond=(NOT p1), acceptance={} source=5 dest: 2}, { cond=p1, acceptance={} source=5 dest: 5}], [{ cond=(AND (NOT p0) (NOT p2)), acceptance={0} source=6 dest: 2}, { cond=(AND p0 (NOT p2)), acceptance={0} source=6 dest: 6}], [{ cond=(AND (NOT p0) (NOT p1) (NOT p2)), acceptance={} source=7 dest: 2}, { cond=(AND (NOT p0) p1 (NOT p2)), acceptance={} source=7 dest: 5}, { cond=(AND p0 (NOT p1) (NOT p2)), acceptance={} source=7 dest: 6}, { cond=(AND p0 p1 (NOT p2)), acceptance={} source=7 dest: 7}]], initial=0, aps=[p0:(LEQ s27 s39), p1:(LEQ 1 s18), p2:(LEQ s2 s18)], nbAcceptance=1, properties=[trans-labels, explicit-labels, trans-acc, no-univ-branch, very-weak, weak, inherently-weak], stateDesc=[null, null, null, null, null, null, null, null][false, false, false, false, false, false, false, false]]
Entered a terminal (fully accepting) state of product in 1 steps with 0 reset in 0 ms.
FORMULA DNAwalker-PT-16redondantChoiceR-07 FALSE TECHNIQUES STUTTER_TEST
Treatment of property DNAwalker-PT-16redondantChoiceR-07 finished in 1100 ms.
Running Spot : CommandLine [args=[/home/mcc/BenchKit/bin//..//ltl2tgba, --hoaf=tv, -f, !(X((G(p0)||X(X(p1)))))], workingDir=/home/mcc/execution]
Support contains 4 out of 43 places. Attempting structural reductions.
Starting structural reductions, iteration 0 : 43/43 places, 490/490 transitions.
Applied a total of 0 rules in 2 ms. Remains 43 /43 variables (removed 0) and now considering 490/490 (removed 0) transitions.
[2021-05-09 08:28:06] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:06] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:06] [INFO ] Dead Transitions using invariants and state equation in 134 ms returned []
[2021-05-09 08:28:06] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:06] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:06] [INFO ] Implicit Places using invariants in 32 ms returned []
[2021-05-09 08:28:06] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:06] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:06] [INFO ] State equation strengthened by 161 read => feed constraints.
[2021-05-09 08:28:06] [INFO ] Implicit Places using invariants and state equation in 528 ms returned []
Implicit Place search using SMT with State Equation took 569 ms to find 0 implicit places.
[2021-05-09 08:28:06] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:06] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:07] [INFO ] Dead Transitions using invariants and state equation in 144 ms returned []
Finished structural reductions, in 1 iterations. Remains : 43/43 places, 490/490 transitions.
Stuttering acceptance computed with spot in 171 ms :[(NOT p1), (AND (NOT p1) (NOT p0)), (AND (NOT p1) (NOT p0)), (AND (NOT p0) (NOT p1)), (NOT p1), true, (AND (NOT p1) (NOT p0)), (NOT p0)]
Running random walk in product with property : DNAwalker-PT-16redondantChoiceR-11 automaton TGBA [mat=[[{ cond=(NOT p1), acceptance={} source=0 dest: 5}], [{ cond=true, acceptance={} source=1 dest: 6}], [{ cond=(NOT p0), acceptance={} source=2 dest: 0}, { cond=p0, acceptance={} source=2 dest: 3}], [{ cond=(AND (NOT p0) (NOT p1)), acceptance={} source=3 dest: 5}, { cond=(AND p0 (NOT p1)), acceptance={} source=3 dest: 7}], [{ cond=true, acceptance={} source=4 dest: 0}], [{ cond=true, acceptance={0} source=5 dest: 5}], [{ cond=p0, acceptance={} source=6 dest: 2}, { cond=(NOT p0), acceptance={} source=6 dest: 4}], [{ cond=(NOT p0), acceptance={} source=7 dest: 5}, { cond=p0, acceptance={} source=7 dest: 7}]], initial=1, aps=[p1:(GT s2 s35), p0:(LEQ s33 s29)], nbAcceptance=1, properties=[trans-labels, explicit-labels, trans-acc, deterministic, no-univ-branch, unambiguous, semi-deterministic, terminal, very-weak, weak, inherently-weak], stateDesc=[null, null, null, null, null, null, null, null][false, false, false, false, false, false, false, false]]
Entered a terminal (fully accepting) state of product in 64 steps with 12 reset in 0 ms.
FORMULA DNAwalker-PT-16redondantChoiceR-11 FALSE TECHNIQUES STUTTER_TEST
Treatment of property DNAwalker-PT-16redondantChoiceR-11 finished in 1042 ms.
Running Spot : CommandLine [args=[/home/mcc/BenchKit/bin//..//ltl2tgba, --hoaf=tv, -f, !(F((p0&&F(p1))))], workingDir=/home/mcc/execution]
Support contains 3 out of 43 places. Attempting structural reductions.
Starting structural reductions, iteration 0 : 43/43 places, 490/490 transitions.
Applied a total of 0 rules in 7 ms. Remains 43 /43 variables (removed 0) and now considering 490/490 (removed 0) transitions.
[2021-05-09 08:28:07] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:07] [INFO ] Computed 0 place invariants in 2 ms
[2021-05-09 08:28:07] [INFO ] Dead Transitions using invariants and state equation in 135 ms returned []
[2021-05-09 08:28:07] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:07] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:07] [INFO ] Implicit Places using invariants in 40 ms returned []
[2021-05-09 08:28:07] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:07] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:07] [INFO ] State equation strengthened by 161 read => feed constraints.
[2021-05-09 08:28:08] [INFO ] Implicit Places using invariants and state equation in 523 ms returned []
Implicit Place search using SMT with State Equation took 564 ms to find 0 implicit places.
[2021-05-09 08:28:08] [INFO ] Redundant transitions in 2 ms returned []
[2021-05-09 08:28:08] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:08] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:08] [INFO ] Dead Transitions using invariants and state equation in 144 ms returned []
Finished structural reductions, in 1 iterations. Remains : 43/43 places, 490/490 transitions.
Stuttering acceptance computed with spot in 51 ms :[(NOT p1), (OR (NOT p0) (NOT p1))]
Running random walk in product with property : DNAwalker-PT-16redondantChoiceR-13 automaton TGBA [mat=[[{ cond=(NOT p1), acceptance={0} source=0 dest: 0}], [{ cond=(AND p0 (NOT p1)), acceptance={0} source=1 dest: 0}, { cond=(NOT p0), acceptance={0} source=1 dest: 1}]], initial=1, aps=[p1:(LEQ s26 s35), p0:(LEQ 1 s19)], nbAcceptance=1, properties=[trans-labels, explicit-labels, trans-acc, colored, deterministic, no-univ-branch, unambiguous, semi-deterministic, stutter-invariant, very-weak, weak, inherently-weak], stateDesc=[null, null][true, true]]
Stuttering criterion allowed to conclude after 16 steps with 0 reset in 0 ms.
FORMULA DNAwalker-PT-16redondantChoiceR-13 FALSE TECHNIQUES STUTTER_TEST
Treatment of property DNAwalker-PT-16redondantChoiceR-13 finished in 928 ms.
Running Spot : CommandLine [args=[/home/mcc/BenchKit/bin//..//ltl2tgba, --hoaf=tv, -f, !(F(G(p0)))], workingDir=/home/mcc/execution]
Support contains 2 out of 43 places. Attempting structural reductions.
Starting structural reductions, iteration 0 : 43/43 places, 490/490 transitions.
Applied a total of 0 rules in 7 ms. Remains 43 /43 variables (removed 0) and now considering 490/490 (removed 0) transitions.
[2021-05-09 08:28:08] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:08] [INFO ] Computed 0 place invariants in 8 ms
[2021-05-09 08:28:08] [INFO ] Dead Transitions using invariants and state equation in 150 ms returned []
[2021-05-09 08:28:08] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:08] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:08] [INFO ] Implicit Places using invariants in 35 ms returned []
[2021-05-09 08:28:08] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:08] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:08] [INFO ] State equation strengthened by 161 read => feed constraints.
[2021-05-09 08:28:09] [INFO ] Implicit Places using invariants and state equation in 542 ms returned []
Implicit Place search using SMT with State Equation took 578 ms to find 0 implicit places.
[2021-05-09 08:28:09] [INFO ] Redundant transitions in 3 ms returned []
[2021-05-09 08:28:09] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:09] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:09] [INFO ] Dead Transitions using invariants and state equation in 145 ms returned []
Finished structural reductions, in 1 iterations. Remains : 43/43 places, 490/490 transitions.
Stuttering acceptance computed with spot in 24 ms :[(NOT p0)]
Running random walk in product with property : DNAwalker-PT-16redondantChoiceR-14 automaton TGBA [mat=[[{ cond=(NOT p0), acceptance={0} source=0 dest: 0}, { cond=p0, acceptance={} source=0 dest: 0}]], initial=0, aps=[p0:(GT s14 s19)], nbAcceptance=1, properties=[trans-labels, explicit-labels, trans-acc, complete, deterministic, no-univ-branch, unambiguous, semi-deterministic, stutter-invariant], stateDesc=[null][true]]
Stuttering criterion allowed to conclude after 10 steps with 0 reset in 0 ms.
FORMULA DNAwalker-PT-16redondantChoiceR-14 FALSE TECHNIQUES STUTTER_TEST
Treatment of property DNAwalker-PT-16redondantChoiceR-14 finished in 1017 ms.
Running Spot : CommandLine [args=[/home/mcc/BenchKit/bin//..//ltl2tgba, --hoaf=tv, -f, !((G(p0) U G((G(p1) U (p2||X((X(p3) U (p2&&X(p3)))))))))], workingDir=/home/mcc/execution]
Support contains 5 out of 43 places. Attempting structural reductions.
Starting structural reductions, iteration 0 : 43/43 places, 490/490 transitions.
Applied a total of 0 rules in 4 ms. Remains 43 /43 variables (removed 0) and now considering 490/490 (removed 0) transitions.
[2021-05-09 08:28:09] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:09] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:09] [INFO ] Dead Transitions using invariants and state equation in 138 ms returned []
[2021-05-09 08:28:09] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:09] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:09] [INFO ] Implicit Places using invariants in 49 ms returned []
[2021-05-09 08:28:09] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:09] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:09] [INFO ] State equation strengthened by 161 read => feed constraints.
[2021-05-09 08:28:10] [INFO ] Implicit Places using invariants and state equation in 563 ms returned []
Implicit Place search using SMT with State Equation took 614 ms to find 0 implicit places.
[2021-05-09 08:28:10] [INFO ] Flow matrix only has 489 transitions (discarded 1 similar events)
// Phase 1: matrix 489 rows 43 cols
[2021-05-09 08:28:10] [INFO ] Computed 0 place invariants in 1 ms
[2021-05-09 08:28:10] [INFO ] Dead Transitions using invariants and state equation in 137 ms returned []
Finished structural reductions, in 1 iterations. Remains : 43/43 places, 490/490 transitions.
Stuttering acceptance computed with spot in 787 ms :[(NOT p2), (OR (NOT p3) (NOT p2)), (NOT p2), (AND (NOT p3) (NOT p0) (NOT p1)), (AND (NOT p1) (NOT p2)), (NOT p2), (OR (AND (NOT p0) p1 (NOT p2)) (AND (NOT p0) (NOT p3))), (NOT p2), (OR (AND (NOT p1) (NOT p3)) (AND (NOT p1) (NOT p2))), (NOT p2), (NOT p3), true, (AND (NOT p0) (NOT p1) (NOT p3)), (AND (NOT p0) (NOT p3)), (AND (NOT p1) (NOT p3)), (AND (NOT p1) (NOT p2) (NOT p3)), (AND (NOT p2) (NOT p3)), (AND (NOT p2) (NOT p1) (NOT p3)), (AND (NOT p2) (NOT p3)), (AND (NOT p2) (NOT p3)), (NOT p0), (NOT p1), (AND (NOT p0) (NOT p1)), (AND (NOT p1) (NOT p2)), (AND (NOT p1) (NOT p2))]
Running random walk in product with property : DNAwalker-PT-16redondantChoiceR-15 automaton TGBA [mat=[[{ cond=p0, acceptance={0} source=0 dest: 0}, { cond=(AND (NOT p2) (NOT p1) (NOT p0)), acceptance={} source=0 dest: 1}, { cond=(NOT p0), acceptance={} source=0 dest: 2}, { cond=(AND (NOT p2) p1 p0), acceptance={} source=0 dest: 3}, { cond=(AND (NOT p2) p1 p0), acceptance={1} source=0 dest: 4}, { cond=(AND (NOT p2) p1 p0), acceptance={0, 1} source=0 dest: 5}, { cond=(AND (NOT p2) (NOT p1) p0), acceptance={} source=0 dest: 6}, { cond=(AND (NOT p2) (NOT p1) p0), acceptance={0, 1} source=0 dest: 7}, { cond=(AND (NOT p2) p1 (NOT p0)), acceptance={} source=0 dest: 8}, { cond=(AND (NOT p2) p1 (NOT p0)), acceptance={} source=0 dest: 9}], [{ cond=(NOT p2), acceptance={0, 1} source=1 dest: 1}, { cond=true, acceptance={} source=1 dest: 10}], [{ cond=(AND (NOT p2) (NOT p1)), acceptance={} source=2 dest: 1}, { cond=true, acceptance={} source=2 dest: 2}, { cond=(AND (NOT p2) p1), acceptance={} source=2 dest: 8}, { cond=(AND (NOT p2) p1), acceptance={} source=2 dest: 9}], [{ cond=(AND (NOT p1) (NOT p0)), acceptance={} source=3 dest: 10}, { cond=(AND p1 p0), acceptance={} source=3 dest: 12}, { cond=(AND (NOT p1) p0), acceptance={} source=3 dest: 13}, { cond=(AND p1 (NOT p0)), acceptance={} source=3 dest: 14}], [{ cond=(AND (NOT p2) (NOT p1) (NOT p0)), acceptance={} source=4 dest: 1}, { cond=(AND (NOT p2) p1 p0), acceptance={} source=4 dest: 3}, { cond=(AND (NOT p2) p1 p0), acceptance={1} source=4 dest: 4}, { cond=(AND (NOT p2) (NOT p1) p0), acceptance={} source=4 dest: 6}, { cond=(AND (NOT p2) (NOT p1) p0), acceptance={0, 1} source=4 dest: 7}, { cond=(AND (NOT p2) p1 (NOT p0)), acceptance={} source=4 dest: 8}, { cond=(AND p1 p0), acceptance={} source=4 dest: 15}, { cond=(AND (NOT p1) p0), acceptance={0} source=4 dest: 16}, { cond=(AND p1 (NOT p0)), acceptance={} source=4 dest: 17}, { cond=(AND (NOT p1) (NOT p0)), acceptance={} source=4 dest: 18}], [{ cond=(AND (NOT p2) (NOT p1) (NOT p0)), acceptance={} source=5 dest: 1}, { cond=(AND (NOT p2) p1 p0), acceptance={} source=5 dest: 3}, { cond=(AND (NOT p2) p1 p0), acceptance={1} source=5 dest: 4}, { cond=(AND (NOT p2) p1 p0), acceptance={0, 1} source=5 dest: 5}, { cond=(AND (NOT p2) (NOT p1) p0), acceptance={} source=5 dest: 6}, { cond=(AND (NOT p2) (NOT p1) p0), acceptance={0, 1} source=5 dest: 7}, { cond=(AND (NOT p2) p1 (NOT p0)), acceptance={} source=5 dest: 8}, { cond=(AND (NOT p2) p1 (NOT p0)), acceptance={} source=5 dest: 9}], [{ cond=(AND (NOT p2) p1 (NOT p0)), acceptance={} source=6 dest: 1}, { cond=(AND (NOT p2) p1 p0), acceptance={} source=6 dest: 6}, { cond=(NOT p0), acceptance={} source=6 dest: 10}, { cond=p0, acceptance={} source=6 dest: 13}], [{ cond=(AND (NOT p2) (NOT p1) (NOT p0)), acceptance={} source=7 dest: 1}, { cond=(AND (NOT p2) p1 p0), acceptance={} source=7 dest: 3}, { cond=(AND (NOT p2) p1 p0), acceptance={1} source=7 dest: 4}, { cond=(AND (NOT p2) p1 p0), acceptance={0, 1} source=7 dest: 5}, { cond=(AND (NOT p2) (NOT p1) p0), acceptance={} source=7 dest: 6}, { cond=(AND (NOT p2) (NOT p1) p0), acceptance={0, 1} source=7 dest: 7}, { cond=(AND (NOT p2) p1 p0), acceptance={0} source=7 dest: 7}, { cond=(AND (NOT p2) p1 (NOT p0)), acceptance={} source=7 dest: 8}, { cond=(AND (NOT p2) p1 (NOT p0)), acceptance={} source=7 dest: 9}, { cond=p0, acceptance={0} source=7 dest: 16}, { cond=(NOT p0), acceptance={} source=7 dest: 18}, { cond=(AND (NOT p2) p1 (NOT p0)), acceptance={} source=7 dest: 19}], [{ cond=(AND (NOT p2) (NOT p1)), acceptance={} source=8 dest: 1}, { cond=(AND (NOT p2) p1), acceptance={} source=8 dest: 8}, { cond=(NOT p1), acceptance={} source=8 dest: 10}, { cond=p1, acceptance={} source=8 dest: 14}], [{ cond=(AND (NOT p2) (NOT p1)), acceptance={} source=9 dest: 1}, { cond=(AND (NOT p2) p1), acceptance={} source=9 dest: 8}, { cond=(AND (NOT p2) p1), acceptance={0, 1} source=9 dest: 9}], [{ cond=(NOT p3), acceptance={} source=10 dest: 11}], [{ cond=true, acceptance={0, 1} source=11 dest: 11}], [{ cond=(AND (NOT p1) (NOT p0) (NOT p3)), acceptance={} source=12 dest: 11}, { cond=(AND (NOT p1) p0 (NOT p3)), acceptance={} source=12 dest: 20}, { cond=(AND p1 (NOT p0) (NOT p3)), acceptance={} source=12 dest: 21}, { cond=(AND p1 p0 (NOT p3)), acceptance={} source=12 dest: 22}], [{ cond=(AND (NOT p0) (NOT p3)), acceptance={} source=13 dest: 11}, { cond=(AND p0 (NOT p3)), acceptance={} source=13 dest: 20}], [{ cond=(AND (NOT p1) (NOT p3)), acceptance={} source=14 dest: 11}, { cond=(AND p1 (NOT p3)), acceptance={} source=14 dest: 21}], [{ cond=(AND (NOT p1) p0 (NOT p3)), acceptance={0} source=15 dest: 0}, { cond=(AND (NOT p2) (NOT p1) (NOT p0) (NOT p3)), acceptance={} source=15 dest: 1}, { cond=(AND (NOT p1) (NOT p0) (NOT p3)), acceptance={} source=15 dest: 2}, { cond=(AND (NOT p2) p1 p0 (NOT p3)), acceptance={} source=15 dest: 3}, { cond=(AND (NOT p2) p1 p0 (NOT p3)), acceptance={1} source=15 dest: 4}, { cond=(AND (NOT p2) (NOT p1) p0 (NOT p3)), acceptance={} source=15 dest: 6}, { cond=(AND (NOT p2) (NOT p1) p0 (NOT p3)), acceptance={0, 1} source=15 dest: 7}, { cond=(AND (NOT p2) p1 (NOT p0) (NOT p3)), acceptance={} source=15 dest: 8}, { cond=(AND p1 (NOT p0) (NOT p3)), acceptance={} source=15 dest: 23}, { cond=(AND p1 p0 (NOT p3)), acceptance={} source=15 dest: 24}], [{ cond=(AND p0 (NOT p3)), acceptance={0} source=16 dest: 0}, { cond=(AND (NOT p2) (NOT p1) (NOT p0) (NOT p3)), acceptance={} source=16 dest: 1}, { cond=(AND (NOT p0) (NOT p3)), acceptance={} source=16 dest: 2}, { cond=(AND (NOT p2) p1 p0 (NOT p3)), acceptance={} source=16 dest: 3}, { cond=(AND (NOT p2) p1 p0 (NOT p3)), acceptance={1} source=16 dest: 4}, { cond=(AND (NOT p2) p1 p0 (NOT p3)), acceptance={0, 1} source=16 dest: 5}, { cond=(AND (NOT p2) (NOT p1) p0 (NOT p3)), acceptance={} source=16 dest: 6}, { cond=(AND (NOT p2) (NOT p1) p0 (NOT p3)), acceptance={0, 1} source=16 dest: 7}, { cond=(AND (NOT p2) p1 (NOT p0) (NOT p3)), acceptance={} source=16 dest: 8}, { cond=(AND (NOT p2) p1 (NOT p0) (NOT p3)), acceptance={} source=16 dest: 9}], [{ cond=(AND (NOT p2) (NOT p1) (NOT p3)), acceptance={} source=17 dest: 1}, { cond=(AND (NOT p1) (NOT p3)), acceptance={} source=17 dest: 2}, { cond=(AND (NOT p2) p1 (NOT p3)), acceptance={} source=17 dest: 8}, { cond=(AND p1 (NOT p3)), acceptance={} source=17 dest: 23}], [{ cond=(AND (NOT p2) (NOT p1) (NOT p3)), acceptance={} source=18 dest: 1}, { cond=(NOT p3), acceptance={} source=18 dest: 2}, { cond=(AND (NOT p2) p1 (NOT p3)), acceptance={} source=18 dest: 8}, { cond=(AND (NOT p2) p1 (NOT p3)), acceptance={} source=18 dest: 9}], [{ cond=true, acceptance={} source=19 dest: 18}, { cond=(AND (NOT p2) p1), acceptance={} source=19 dest: 19}], [{ cond=(NOT p0), acceptance={} source=20 dest: 11}, { cond=p0, acceptance={} source=20 dest: 20}], [{ cond=(NOT p1), acceptance={} source=21 dest: 11}, { cond=p1, acceptance={} source=21 dest: 21}], [{ cond=(AND (NOT p1) (NOT p0)), acceptance={} source=22 dest: 11}, { cond=(AND (NOT p1) p0), acceptance={} source=22 dest: 20}, { cond=(AND p1 (NOT p0)), acceptance={} source=22 dest: 21}, { cond=(AND p1 p0), acceptance={} source=22 dest: 22}], [{ cond=(AND (NOT p2) (NOT p1)), acceptance={} source=23 dest: 1}, { cond=(NOT p1), acceptance={} source=23 dest: 2}, { cond=(AND (NOT p2) p1), acceptance={} source=23 dest: 8}, { cond=p1, acceptance={} source=23 dest: 23}], [{ cond=(AND (NOT p1) p0), acceptance={0} source=24 dest: 0}, { cond=(AND (NOT p2) (NOT p1) (NOT p0)), acceptance={} source=24 dest: 1}, { cond=(AND (NOT p1) (NOT p0)), acceptance={} source=24 dest: 2}, { cond=(AND (NOT p2) p1 p0), acceptance={} source=24 dest: 3}, { cond=(AND (NOT p2) p1 p0), acceptance={1} source=24 dest: 4}, { cond=(AND (NOT p2) (NOT p1) p0), acceptance={} source=24 dest: 6}, { cond=(AND (NOT p2) (NOT p1) p0), acceptance={0, 1} source=24 dest: 7}, { cond=(AND (NOT p2) p1 (NOT p0)), acceptance={} source=24 dest: 8}, { cond=(AND p1 (NOT p0)), acceptance={} source=24 dest: 23}, { cond=(AND p1 p0), acceptance={} source=24 dest: 24}]], initial=0, aps=[p0:(LEQ s42 s3), p2:(LEQ 2 s4), p1:(LEQ 1 s12), p3:(LEQ 1 s34)], nbAcceptance=2, properties=[trans-labels, explicit-labels, trans-acc, no-univ-branch], stateDesc=[null, null, null, null, null, null, null, null, null, null, null, null, null, null, null, null, null, null, null, null, null, null, null, null, null][false, false, false, false, false, false, false, false, false, false, false, false, false, false, false, false, false, false, false, false, false, false, false, false, false]]
Entered a terminal (fully accepting) state of product in 26 steps with 0 reset in 0 ms.
FORMULA DNAwalker-PT-16redondantChoiceR-15 FALSE TECHNIQUES STUTTER_TEST
Treatment of property DNAwalker-PT-16redondantChoiceR-15 finished in 1711 ms.
Using solver Z3 to compute partial order matrices.
Built C files in :
/tmp/ltsmin10744474714391721522
[2021-05-09 08:28:10] [INFO ] Computing symmetric may disable matrix : 490 transitions.
[2021-05-09 08:28:10] [INFO ] Computation of Complete disable matrix. took 4 ms. Total solver calls (SAT/UNSAT): 0(0/0)
[2021-05-09 08:28:10] [INFO ] Computing symmetric may enable matrix : 490 transitions.
[2021-05-09 08:28:10] [INFO ] Applying decomposition
[2021-05-09 08:28:10] [INFO ] Flatten gal took : 14 ms
[2021-05-09 08:28:10] [INFO ] Computation of Complete enable matrix. took 15 ms. Total solver calls (SAT/UNSAT): 0(0/0)
Converted graph to binary with : CommandLine [args=[/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.louvain.binaries_1.0.0.202104292328/bin/convert-linux64, -i, /tmp/graph1195713272845829759.txt, -o, /tmp/graph1195713272845829759.bin, -w, /tmp/graph1195713272845829759.weights], workingDir=null]
Built communities with : CommandLine [args=[/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.louvain.binaries_1.0.0.202104292328/bin/louvain-linux64, /tmp/graph1195713272845829759.bin, -l, -1, -v, -w, /tmp/graph1195713272845829759.weights, -q, 0, -e, 0.001], workingDir=null]
[2021-05-09 08:28:11] [INFO ] Decomposing Gal with order
[2021-05-09 08:28:11] [INFO ] Rewriting arrays to variables to allow decomposition.
[2021-05-09 08:28:11] [INFO ] Removed a total of 868 redundant transitions.
[2021-05-09 08:28:11] [INFO ] Computing Do-Not-Accords matrix : 490 transitions.
[2021-05-09 08:28:11] [INFO ] Flatten gal took : 44 ms
[2021-05-09 08:28:11] [INFO ] Fuse similar labels procedure discarded/fused a total of 0 labels/synchronizations in 2 ms.
[2021-05-09 08:28:11] [INFO ] Computation of Completed DNA matrix. took 16 ms. Total solver calls (SAT/UNSAT): 0(0/0)
[2021-05-09 08:28:11] [INFO ] Built C files in 135ms conformant to PINS (ltsmin variant)in folder :/tmp/ltsmin10744474714391721522
Running compilation step : CommandLine [args=[gcc, -c, -I/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.ltsmin.binaries_1.0.0.202104292328/bin/include/, -I., -std=c99, -fPIC, -O2, model.c], workingDir=/tmp/ltsmin10744474714391721522]
[2021-05-09 08:28:11] [INFO ] Time to serialize gal into /tmp/LTLCardinality5352829005014878488.gal : 6 ms
[2021-05-09 08:28:11] [INFO ] Time to serialize properties into /tmp/LTLCardinality13899439673695382927.ltl : 1 ms
Invoking ITS tools like this :CommandLine [args=[/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.itstools.binaries_1.0.0.202104292328/bin/its-ltl-linux64, --gc-threshold, 2000000, -i, /tmp/LTLCardinality5352829005014878488.gal, -t, CGAL, -LTL, /tmp/LTLCardinality13899439673695382927.ltl, -c, -stutter-deadlock, --gen-order, FOLLOW], workingDir=/home/mcc/execution]
its-ltl command run as :
/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.itstools.binaries_1.0.0.202104292328/bin/its-ltl-linux64 --gc-threshold 2000000 -i /tmp/LTLCardinality5352829005014878488.gal -t CGAL -LTL /tmp/LTLCardinality13899439673695382927.ltl -c -stutter-deadlock --gen-order FOLLOW
Read 2 LTL properties
Checking formula 0 : !(((F(!("(((((gu0.A15<=gu0.A1)&&(gu13.A16<1))&&(gu21.A24>=1))||(gu21.A24>=1))&&(gu0.A15>gu0.A1))")))U((("(gu0.A15>gu0.A1)")&&(F(!("(((((gu0.A15<=gu0.A1)&&(gu13.A16<1))&&(gu21.A24>=1))||(gu21.A24>=1))&&(gu0.A15>gu0.A1))"))))||(G(F(!("(((((gu0.A15<=gu0.A1)&&(gu13.A16<1))&&(gu21.A24>=1))||(gu21.A24>=1))&&(gu0.A15>gu0.A1))")))))))
Formula 0 simplified : !(F!"(((((gu0.A15<=gu0.A1)&&(gu13.A16<1))&&(gu21.A24>=1))||(gu21.A24>=1))&&(gu0.A15>gu0.A1))" U (("(gu0.A15>gu0.A1)" & F!"(((((gu0.A15<=gu0.A1)&&(gu13.A16<1))&&(gu21.A24>=1))||(gu21.A24>=1))&&(gu0.A15>gu0.A1))") | GF!"(((((gu0.A15<=gu0.A1)&&(gu13.A16<1))&&(gu21.A24>=1))||(gu21.A24>=1))&&(gu0.A15>gu0.A1))"))
Compilation finished in 2814 ms.
Running link step : CommandLine [args=[gcc, -shared, -o, gal.so, model.o], workingDir=/tmp/ltsmin10744474714391721522]
Link finished in 57 ms.
Running LTSmin : CommandLine [args=[/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.ltsmin.binaries_1.0.0.202104292328/bin/pins2lts-mc-linux64, ./gal.so, --threads=8, -p, --pins-guards, --when, --ltl, (<>((LTLAPp0==true)) U ([](<>((LTLAPp1==true)))||((LTLAPp2==true)&&<>((LTLAPp1==true))))), --buchi-type=spotba], workingDir=/tmp/ltsmin10744474714391721522]
LTSmin run took 872 ms.
FORMULA DNAwalker-PT-16redondantChoiceR-01 FALSE TECHNIQUES PARTIAL_ORDER EXPLICIT LTSMIN SAT_SMT
Running LTSmin : CommandLine [args=[/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.ltsmin.binaries_1.0.0.202104292328/bin/pins2lts-mc-linux64, ./gal.so, --threads=8, -p, --pins-guards, --when, --ltl, ((LTLAPp3==true) U []((LTLAPp4==true))), --buchi-type=spotba], workingDir=/tmp/ltsmin10744474714391721522]
LTSmin run took 862 ms.
FORMULA DNAwalker-PT-16redondantChoiceR-02 FALSE TECHNIQUES PARTIAL_ORDER EXPLICIT LTSMIN SAT_SMT
ITS tools runner thread asked to quit. Dying gracefully.
BK_STOP 1620548896724
--------------------
content from stderr:
+ export BINDIR=/home/mcc/BenchKit/bin//../
+ BINDIR=/home/mcc/BenchKit/bin//../
++ pwd
+ export MODEL=/home/mcc/execution
+ MODEL=/home/mcc/execution
+ [[ LTLCardinality = StateSpace ]]
+ /home/mcc/BenchKit/bin//..//runeclipse.sh /home/mcc/execution LTLCardinality -its -ltsmin -greatspnpath /home/mcc/BenchKit/bin//..//greatspn/ -order META -manyOrder -smt -timeout 3600
+ ulimit -s 65536
+ [[ -z '' ]]
+ export LTSMIN_MEM_SIZE=8589934592
+ LTSMIN_MEM_SIZE=8589934592
++ cut -d . -f 9
++ ls /home/mcc/BenchKit/bin//..//itstools/plugins/fr.lip6.move.gal.application.pnmcc_1.0.0.202104292328.jar
+ VERSION=0
+ echo 'Running Version 0'
+ /home/mcc/BenchKit/bin//..//itstools/its-tools -data /home/mcc/execution/workspace -pnfolder /home/mcc/execution -examination LTLCardinality -spotpath /home/mcc/BenchKit/bin//..//ltlfilt -z3path /home/mcc/BenchKit/bin//..//z3/bin/z3 -yices2path /home/mcc/BenchKit/bin//..//yices/bin/yices -its -ltsmin -greatspnpath /home/mcc/BenchKit/bin//..//greatspn/ -order META -manyOrder -smt -timeout 3600 -vmargs -Dosgi.locking=none -Declipse.stateSaveDelayInterval=-1 -Dosgi.configuration.area=/tmp/.eclipse -Xss128m -Xms40m -Xmx16000m
Sequence of Actions to be Executed by the VM
This is useful if one wants to reexecute the tool in the VM from the submitted image disk.
set -x
# this is for BenchKit: configuration of major elements for the test
export BK_INPUT="DNAwalker-PT-16redondantChoiceR"
export BK_EXAMINATION="LTLCardinality"
export BK_TOOL="itstools"
export BK_RESULT_DIR="/tmp/BK_RESULTS/OUTPUTS"
export BK_TIME_CONFINEMENT="3600"
export BK_MEMORY_CONFINEMENT="16384"
export BK_BIN_PATH="/home/mcc/BenchKit/bin/"
# this is specific to your benchmark or test
export BIN_DIR="$HOME/BenchKit/bin"
# remove the execution directoty if it exists (to avoid increse of .vmdk images)
if [ -d execution ] ; then
rm -rf execution
fi
# this is for BenchKit: explicit launching of the test
echo "====================================================================="
echo " Generated by BenchKit 2-4028"
echo " Executing tool itstools"
echo " Input is DNAwalker-PT-16redondantChoiceR, examination is LTLCardinality"
echo " Time confinement is $BK_TIME_CONFINEMENT seconds"
echo " Memory confinement is 16384 MBytes"
echo " Number of cores is 4"
echo " Run identifier is r064-tall-162038397500652"
echo "====================================================================="
echo
echo "--------------------"
echo "preparation of the directory to be used:"
tar xzf /home/mcc/BenchKit/INPUTS/DNAwalker-PT-16redondantChoiceR.tgz
mv DNAwalker-PT-16redondantChoiceR execution
cd execution
if [ "LTLCardinality" = "ReachabilityDeadlock" ] || [ "LTLCardinality" = "UpperBounds" ] || [ "LTLCardinality" = "QuasiLiveness" ] || [ "LTLCardinality" = "StableMarking" ] || [ "LTLCardinality" = "Liveness" ] || [ "LTLCardinality" = "OneSafe" ] || [ "LTLCardinality" = "StateSpace" ]; then
rm -f GenericPropertiesVerdict.xml
fi
pwd
ls -lh
echo
echo "--------------------"
echo "content from stdout:"
echo
echo "=== Data for post analysis generated by BenchKit (invocation template)"
echo
if [ "LTLCardinality" = "UpperBounds" ] ; then
echo "The expected result is a vector of positive values"
echo NUM_VECTOR
elif [ "LTLCardinality" != "StateSpace" ] ; then
echo "The expected result is a vector of booleans"
echo BOOL_VECTOR
else
echo "no data necessary for post analysis"
fi
echo
if [ -f "LTLCardinality.txt" ] ; then
echo "here is the order used to build the result vector(from text file)"
for x in $(grep Property LTLCardinality.txt | cut -d ' ' -f 2 | sort -u) ; do
echo "FORMULA_NAME $x"
done
elif [ -f "LTLCardinality.xml" ] ; then # for cunf (txt files deleted;-)
echo echo "here is the order used to build the result vector(from xml file)"
for x in $(grep '
echo "FORMULA_NAME $x"
done
elif [ "LTLCardinality" = "ReachabilityDeadlock" ] || [ "LTLCardinality" = "QuasiLiveness" ] || [ "LTLCardinality" = "StableMarking" ] || [ "LTLCardinality" = "Liveness" ] || [ "LTLCardinality" = "OneSafe" ] ; then
echo "FORMULA_NAME LTLCardinality"
fi
echo
echo "=== Now, execution of the tool begins"
echo
echo -n "BK_START "
date -u +%s%3N
echo
timeout -s 9 $BK_TIME_CONFINEMENT bash -c "/home/mcc/BenchKit/BenchKit_head.sh 2> STDERR ; echo ; echo -n \"BK_STOP \" ; date -u +%s%3N"
if [ $? -eq 137 ] ; then
echo
echo "BK_TIME_CONFINEMENT_REACHED"
fi
echo
echo "--------------------"
echo "content from stderr:"
echo
cat STDERR ;

