About the Execution of Tapaal for DNAwalker-PT-05track28LR
| Execution Summary | |||||
| Max Memory Used (MB) |
Time wait (ms) | CPU Usage (ms) | I/O Wait (ms) | Computed Result | Execution Status |
| 2226.331 | 1716100.00 | 6263513.00 | 177.40 | 2 1 2 2 2 2 2 2 2 2 2 2 1 2 1 1 | normal |
Execution Chart
We display below the execution chart for this examination (boot time has been removed).
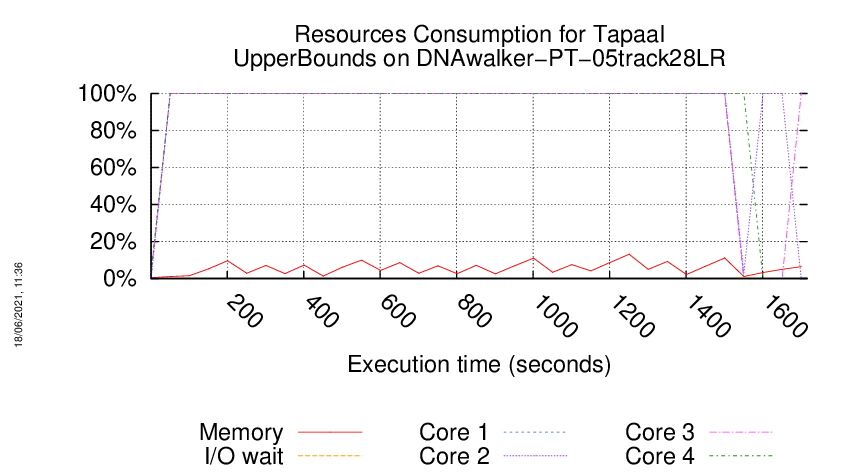
Trace from the execution
Formatting '/data/fkordon/mcc2021-input.r062-tall-162038394700563.qcow2', fmt=qcow2 size=4294967296 backing_file='/data/fkordon/mcc2021-input.qcow2' encryption=off cluster_size=65536 lazy_refcounts=off
Waiting for the VM to be ready (probing ssh)
..............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................
=====================================================================
Generated by BenchKit 2-4028
Executing tool tapaal
Input is DNAwalker-PT-05track28LR, examination is UpperBounds
Time confinement is 3600 seconds
Memory confinement is 16384 MBytes
Number of cores is 4
Run identifier is r062-tall-162038394700563
=====================================================================
--------------------
preparation of the directory to be used:
/home/mcc/execution
total 504K
-rw-r--r-- 1 mcc users 14K May 5 16:58 CTLCardinality.txt
-rw-r--r-- 1 mcc users 144K May 5 16:58 CTLCardinality.xml
-rw-r--r-- 1 mcc users 12K May 5 16:58 CTLFireability.txt
-rw-r--r-- 1 mcc users 92K May 5 16:58 CTLFireability.xml
-rw-r--r-- 1 mcc users 4.2K May 6 14:48 GenericPropertiesDefinition.xml
-rw-r--r-- 1 mcc users 3.4K Mar 28 16:00 LTLCardinality.txt
-rw-r--r-- 1 mcc users 26K Mar 28 16:00 LTLCardinality.xml
-rw-r--r-- 1 mcc users 2.1K Mar 28 16:00 LTLFireability.txt
-rw-r--r-- 1 mcc users 17K Mar 28 16:00 LTLFireability.xml
-rw-r--r-- 1 mcc users 3.7K Mar 23 10:01 ReachabilityCardinality.txt
-rw-r--r-- 1 mcc users 19K Mar 23 10:01 ReachabilityCardinality.xml
-rw-r--r-- 1 mcc users 3.2K Mar 22 17:36 ReachabilityFireability.txt
-rw-r--r-- 1 mcc users 20K Mar 22 17:36 ReachabilityFireability.xml
-rw-r--r-- 1 mcc users 1.7K Mar 22 09:10 UpperBounds.txt
-rw-r--r-- 1 mcc users 3.7K Mar 22 09:10 UpperBounds.xml
-rw-r--r-- 1 mcc users 6 May 5 16:51 equiv_col
-rw-r--r-- 1 mcc users 12 May 5 16:51 instance
-rw-r--r-- 1 mcc users 6 May 5 16:51 iscolored
-rw-r--r-- 1 mcc users 108K May 5 16:51 model.pnml
--------------------
content from stdout:
=== Data for post analysis generated by BenchKit (invocation template)
The expected result is a vector of positive values
NUM_VECTOR
here is the order used to build the result vector(from text file)
FORMULA_NAME DNAwalker-PT-05track28LR-UpperBounds-00
FORMULA_NAME DNAwalker-PT-05track28LR-UpperBounds-01
FORMULA_NAME DNAwalker-PT-05track28LR-UpperBounds-02
FORMULA_NAME DNAwalker-PT-05track28LR-UpperBounds-03
FORMULA_NAME DNAwalker-PT-05track28LR-UpperBounds-04
FORMULA_NAME DNAwalker-PT-05track28LR-UpperBounds-05
FORMULA_NAME DNAwalker-PT-05track28LR-UpperBounds-06
FORMULA_NAME DNAwalker-PT-05track28LR-UpperBounds-07
FORMULA_NAME DNAwalker-PT-05track28LR-UpperBounds-08
FORMULA_NAME DNAwalker-PT-05track28LR-UpperBounds-09
FORMULA_NAME DNAwalker-PT-05track28LR-UpperBounds-10
FORMULA_NAME DNAwalker-PT-05track28LR-UpperBounds-11
FORMULA_NAME DNAwalker-PT-05track28LR-UpperBounds-12
FORMULA_NAME DNAwalker-PT-05track28LR-UpperBounds-13
FORMULA_NAME DNAwalker-PT-05track28LR-UpperBounds-14
FORMULA_NAME DNAwalker-PT-05track28LR-UpperBounds-15
=== Now, execution of the tool begins
BK_START 1620501840882
tapaal
Got BK_BIN_PATH=/home/mcc/BenchKit/bin/
---> tapaal --- TAPAAL
Setting MODEL_PATH=.
Setting VERIFYPN=/home/mcc/BenchKit/bin/verifypn
Got BK_TIME_CONFINEMENT=3600
Setting TEMPDIR=/home/mcc/BenchKit/bin/tmp
Got BK_MEMORY_CONFINEMENT=16384
Limiting to 16265216 kB
Total timeout: 3590
Time left: 3590
*****************************************
* TAPAAL CLASSIC verifying UpperBounds *
*****************************************
/home/mcc/BenchKit/bin/tmp
/home/mcc/BenchKit/bin/tmp/tmp.KzRBvI1Kih
/home/mcc/BenchKit/bin/tmp/tmp.8PkGA2orgB
Time left: 3590
---------------------------------------------------
Step -1: Stripping Colors
---------------------------------------------------
Verifying stripped models (16 in total)
Time left: 3590
---------------------------------------------------
Step 0: Parallel Simplification
---------------------------------------------------
Doing parallel simplification (16 in total)
Total simplification timout is 718 -- reduction timeout is 299
timeout 3590 /home/mcc/BenchKit/bin/verifypn -n -q 718 -l 29 -d 299 -z 4 -s OverApprox --binary-query-io 2 --write-simplified /home/mcc/BenchKit/bin/tmp/tmp.KzRBvI1Kih --write-reduced /home/mcc/BenchKit/bin/tmp/tmp.8PkGA2orgB -x 1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16 ./model.pnml ./UpperBounds.xml
Time left: 3503
---------------------------------------------------
Step 1: Parallel processing
---------------------------------------------------
Doing parallel verification of individual queries (16 in total)
Each query is verified by 4 parallel strategies for 138 seconds
------------------- QUERY 1 ----------------------
No solution found
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Time left: 3361
------------------- QUERY 2 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
@@@0.01,6764@@@
Query index 0 was solved
Query is satisfied.
Search strategy option was ignored as the TAR engine is called.
FORMULA DNAwalker-PT-05track28LR-UpperBounds-01 1 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT TRACE_ABSTRACTION_REFINEMENT
Query index 0 was solved
Query is satisfied.
@@@0.02,6936@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.8PkGA2orgB /home/mcc/BenchKit/bin/tmp/tmp.KzRBvI1Kih --binary-query-io 1 -x 2 -n
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -p\ -s\ DFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.8PkGA2orgB /home/mcc/BenchKit/bin/tmp/tmp.KzRBvI1Kih --binary-query-io 1 -x 2 -n
FORMULA DNAwalker-PT-05track28LR-UpperBounds-01 1 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION
Time left: 3361
------------------- QUERY 3 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is NOT satisfied.
@@@102.18,465276@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.8PkGA2orgB /home/mcc/BenchKit/bin/tmp/tmp.KzRBvI1Kih --binary-query-io 1 -x 3 -n
FORMULA DNAwalker-PT-05track28LR-UpperBounds-02 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3259
------------------- QUERY 4 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is NOT satisfied.
@@@111.56,496116@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.8PkGA2orgB /home/mcc/BenchKit/bin/tmp/tmp.KzRBvI1Kih --binary-query-io 1 -x 4 -n
FORMULA DNAwalker-PT-05track28LR-UpperBounds-03 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3147
------------------- QUERY 5 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is NOT satisfied.
@@@119.43,540620@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.8PkGA2orgB /home/mcc/BenchKit/bin/tmp/tmp.KzRBvI1Kih --binary-query-io 1 -x 5 -n
FORMULA DNAwalker-PT-05track28LR-UpperBounds-04 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 3027
------------------- QUERY 6 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is NOT satisfied.
@@@116.22,527132@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.8PkGA2orgB /home/mcc/BenchKit/bin/tmp/tmp.KzRBvI1Kih --binary-query-io 1 -x 6 -n
FORMULA DNAwalker-PT-05track28LR-UpperBounds-05 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 2911
------------------- QUERY 7 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is NOT satisfied.
@@@103.53,438632@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.8PkGA2orgB /home/mcc/BenchKit/bin/tmp/tmp.KzRBvI1Kih --binary-query-io 1 -x 7 -n
FORMULA DNAwalker-PT-05track28LR-UpperBounds-06 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 2807
------------------- QUERY 8 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is NOT satisfied.
@@@99.04,461392@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.8PkGA2orgB /home/mcc/BenchKit/bin/tmp/tmp.KzRBvI1Kih --binary-query-io 1 -x 8 -n
FORMULA DNAwalker-PT-05track28LR-UpperBounds-07 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 2708
------------------- QUERY 9 ----------------------
No solution found
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Time left: 2567
------------------- QUERY 10 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is NOT satisfied.
@@@94.01,421596@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.8PkGA2orgB /home/mcc/BenchKit/bin/tmp/tmp.KzRBvI1Kih --binary-query-io 1 -x 10 -n
FORMULA DNAwalker-PT-05track28LR-UpperBounds-09 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 2473
------------------- QUERY 11 ----------------------
No solution found
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Time left: 2333
------------------- QUERY 12 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is NOT satisfied.
@@@127.35,530056@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.8PkGA2orgB /home/mcc/BenchKit/bin/tmp/tmp.KzRBvI1Kih --binary-query-io 1 -x 12 -n
FORMULA DNAwalker-PT-05track28LR-UpperBounds-11 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 2205
------------------- QUERY 13 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
@@@0.00,7052@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.8PkGA2orgB /home/mcc/BenchKit/bin/tmp/tmp.KzRBvI1Kih --binary-query-io 1 -x 13 -n
FORMULA DNAwalker-PT-05track28LR-UpperBounds-12 1 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 2205
------------------- QUERY 14 ----------------------
No solution found
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Time left: 2065
------------------- QUERY 15 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
@@@0.00,7016@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -p\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.8PkGA2orgB /home/mcc/BenchKit/bin/tmp/tmp.KzRBvI1Kih --binary-query-io 1 -x 15 -n
FORMULA DNAwalker-PT-05track28LR-UpperBounds-14 1 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION
Time left: 2065
------------------- QUERY 16 ----------------------
Solution found by parallel processing (step 1)
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "en_US.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
Query index 0 was solved
Query is satisfied.
@@@0.00,6808@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -s\ BFS\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.8PkGA2orgB /home/mcc/BenchKit/bin/tmp/tmp.KzRBvI1Kih --binary-query-io 1 -x 16 -n
FORMULA DNAwalker-PT-05track28LR-UpperBounds-15 1 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION
Query index 0 was solved
Query is satisfied.
@@@0.00,6932@@@
parallel: This job succeeded:
eval /usr/bin/time -f "@@@%e,%M@@@" /home/mcc/BenchKit/bin/verifypn -n -n\ -p\ -q\ 0\ -l\ 0\ -d\ 119 /home/mcc/BenchKit/bin/tmp/tmp.8PkGA2orgB /home/mcc/BenchKit/bin/tmp/tmp.KzRBvI1Kih --binary-query-io 1 -x 16 -n
FORMULA DNAwalker-PT-05track28LR-UpperBounds-15 1 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION STUBBORN_SETS
Time left: 2065
---------------------------------------------------
Step 2: Sequential processing
---------------------------------------------------
Remaining 4 queries are verified sequentially.
Each query is verified for a dynamic timeout (at least 0 seconds)
Time left: 2065
Time left: 2065
---------------------------------------------------
Step 3: Multiquery processing
---------------------------------------------------
Remaining 4 queries are solved using multiquery
Time remaining: 2065 seconds of the initial 3590 seconds
Running multiquery on -x 1,9,11,14 for 1548 seconds
FORMULA DNAwalker-PT-05track28LR-UpperBounds-00 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION
Query index 0 was solved
Query is NOT satisfied.
FORMULA DNAwalker-PT-05track28LR-UpperBounds-08 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION
Query index 1 was solved
Query is NOT satisfied.
FORMULA DNAwalker-PT-05track28LR-UpperBounds-10 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION
Query index 2 was solved
Query is NOT satisfied.
FORMULA DNAwalker-PT-05track28LR-UpperBounds-13 2 TECHNIQUES COLLATERAL_PROCESSING STRUCTURAL_REDUCTION QUERY_REDUCTION SAT_SMT EXPLICIT STATE_COMPRESSION
Query index 3 was solved
Query is NOT satisfied.
Solution found by multiquery processing (step 3) for query index 3
Solution found by multiquery processing (step 3) for query index 2
Solution found by multiquery processing (step 3) for query index 1
Solution found by multiquery processing (step 3) for query index 0
Time left: 1874
All queries are solved
Time left: 1874
BK_STOP 1620503556982
--------------------
content from stderr:
CPN OverApproximation is only usable on colored models
glpk mip: timeout
glpk mip: timeout
glpk mip: timeout
glpk mip: timeout
glpk mip: timeout
glpk mip: timeout
glpk mip: timeout
glpk mip: timeout
glpk mip: timeout
glpk mip: timeout
glpk mip: timeout
glpk mip: timeout
glpk mip: timeout
glpk mip: timeout
glpk mip: timeout
glpk mip: timeout
"@@@191.28,1080164@@@"
Sequence of Actions to be Executed by the VM
This is useful if one wants to reexecute the tool in the VM from the submitted image disk.
set -x
# this is for BenchKit: configuration of major elements for the test
export BK_INPUT="DNAwalker-PT-05track28LR"
export BK_EXAMINATION="UpperBounds"
export BK_TOOL="tapaal"
export BK_RESULT_DIR="/tmp/BK_RESULTS/OUTPUTS"
export BK_TIME_CONFINEMENT="3600"
export BK_MEMORY_CONFINEMENT="16384"
export BK_BIN_PATH="/home/mcc/BenchKit/bin/"
# this is specific to your benchmark or test
export BIN_DIR="$HOME/BenchKit/bin"
# remove the execution directoty if it exists (to avoid increse of .vmdk images)
if [ -d execution ] ; then
rm -rf execution
fi
# this is for BenchKit: explicit launching of the test
echo "====================================================================="
echo " Generated by BenchKit 2-4028"
echo " Executing tool tapaal"
echo " Input is DNAwalker-PT-05track28LR, examination is UpperBounds"
echo " Time confinement is $BK_TIME_CONFINEMENT seconds"
echo " Memory confinement is 16384 MBytes"
echo " Number of cores is 4"
echo " Run identifier is r062-tall-162038394700563"
echo "====================================================================="
echo
echo "--------------------"
echo "preparation of the directory to be used:"
tar xzf /home/mcc/BenchKit/INPUTS/DNAwalker-PT-05track28LR.tgz
mv DNAwalker-PT-05track28LR execution
cd execution
if [ "UpperBounds" = "ReachabilityDeadlock" ] || [ "UpperBounds" = "UpperBounds" ] || [ "UpperBounds" = "QuasiLiveness" ] || [ "UpperBounds" = "StableMarking" ] || [ "UpperBounds" = "Liveness" ] || [ "UpperBounds" = "OneSafe" ] || [ "UpperBounds" = "StateSpace" ]; then
rm -f GenericPropertiesVerdict.xml
fi
pwd
ls -lh
echo
echo "--------------------"
echo "content from stdout:"
echo
echo "=== Data for post analysis generated by BenchKit (invocation template)"
echo
if [ "UpperBounds" = "UpperBounds" ] ; then
echo "The expected result is a vector of positive values"
echo NUM_VECTOR
elif [ "UpperBounds" != "StateSpace" ] ; then
echo "The expected result is a vector of booleans"
echo BOOL_VECTOR
else
echo "no data necessary for post analysis"
fi
echo
if [ -f "UpperBounds.txt" ] ; then
echo "here is the order used to build the result vector(from text file)"
for x in $(grep Property UpperBounds.txt | cut -d ' ' -f 2 | sort -u) ; do
echo "FORMULA_NAME $x"
done
elif [ -f "UpperBounds.xml" ] ; then # for cunf (txt files deleted;-)
echo echo "here is the order used to build the result vector(from xml file)"
for x in $(grep '
echo "FORMULA_NAME $x"
done
elif [ "UpperBounds" = "ReachabilityDeadlock" ] || [ "UpperBounds" = "QuasiLiveness" ] || [ "UpperBounds" = "StableMarking" ] || [ "UpperBounds" = "Liveness" ] || [ "UpperBounds" = "OneSafe" ] ; then
echo "FORMULA_NAME UpperBounds"
fi
echo
echo "=== Now, execution of the tool begins"
echo
echo -n "BK_START "
date -u +%s%3N
echo
timeout -s 9 $BK_TIME_CONFINEMENT bash -c "/home/mcc/BenchKit/BenchKit_head.sh 2> STDERR ; echo ; echo -n \"BK_STOP \" ; date -u +%s%3N"
if [ $? -eq 137 ] ; then
echo
echo "BK_TIME_CONFINEMENT_REACHED"
fi
echo
echo "--------------------"
echo "content from stderr:"
echo
cat STDERR ;

