About the Execution of GreatSPN for DNAwalker-PT-16redondantChoiceR
| Execution Summary | |||||
| Max Memory Used (MB) |
Time wait (ms) | CPU Usage (ms) | I/O Wait (ms) | Computed Result | Execution Status |
| 15750.690 | 1056858.00 | 2577898.00 | 111.10 | FFFTFFTFFFTFFFFF | normal |
Execution Chart
We display below the execution chart for this examination (boot time has been removed).
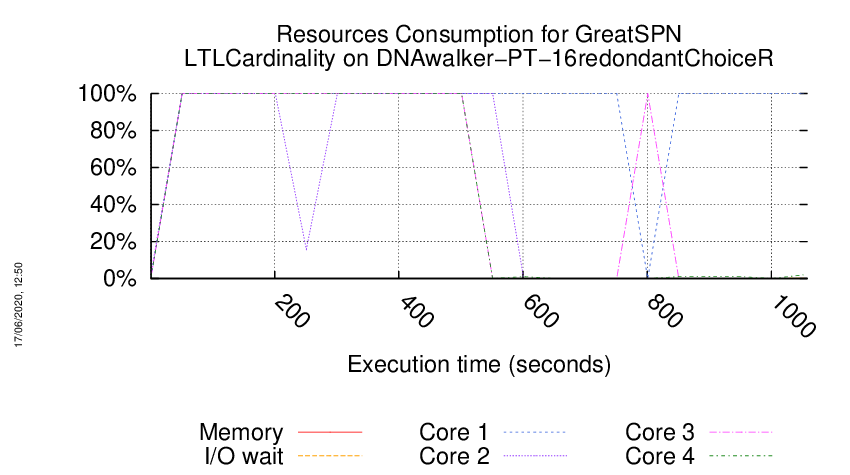
Trace from the execution
Formatting '/mnt/tpsp/fkordon/mcc2020-input.r065-tajo-158922812400201.qcow2', fmt=qcow2 size=4294967296 backing_file='/mnt/tpsp/fkordon/mcc2020-input.qcow2' encryption=off cluster_size=65536 lazy_refcounts=off
Waiting for the VM to be ready (probing ssh)
...........................................................................................................................................................................................................................................................................................................................................................................
=====================================================================
Generated by BenchKit 2-4028
Executing tool gspn
Input is DNAwalker-PT-16redondantChoiceR, examination is LTLCardinality
Time confinement is 3600 seconds
Memory confinement is 16384 MBytes
Number of cores is 4
Run identifier is r065-tajo-158922812400201
=====================================================================
--------------------
preparation of the directory to be used:
/home/mcc/execution
total 396K
-rw-r--r-- 1 mcc users 3.9K Mar 31 05:00 CTLCardinality.txt
-rw-r--r-- 1 mcc users 22K Mar 31 05:00 CTLCardinality.xml
-rw-r--r-- 1 mcc users 3.0K Mar 29 20:56 CTLFireability.txt
-rw-r--r-- 1 mcc users 19K Mar 29 20:56 CTLFireability.xml
-rw-r--r-- 1 mcc users 4.0K Mar 24 05:37 GenericPropertiesDefinition.xml
-rw-r--r-- 1 mcc users 5.6K Mar 24 05:37 GenericPropertiesVerdict.xml
-rw-r--r-- 1 mcc users 3.3K Apr 8 14:52 LTLCardinality.txt
-rw-r--r-- 1 mcc users 23K Apr 28 14:00 LTLCardinality.xml
-rw-r--r-- 1 mcc users 2.2K Apr 8 14:52 LTLFireability.txt
-rw-r--r-- 1 mcc users 15K Apr 28 14:00 LTLFireability.xml
-rw-r--r-- 1 mcc users 4.0K Mar 28 13:43 ReachabilityCardinality.txt
-rw-r--r-- 1 mcc users 21K Mar 28 13:43 ReachabilityCardinality.xml
-rw-r--r-- 1 mcc users 3.3K Mar 27 06:27 ReachabilityFireability.txt
-rw-r--r-- 1 mcc users 20K Mar 27 06:27 ReachabilityFireability.xml
-rw-r--r-- 1 mcc users 1.8K Mar 28 14:49 UpperBounds.txt
-rw-r--r-- 1 mcc users 3.9K Mar 28 14:49 UpperBounds.xml
-rw-r--r-- 1 mcc users 6 Mar 24 05:37 equiv_col
-rw-r--r-- 1 mcc users 19 Mar 24 05:37 instance
-rw-r--r-- 1 mcc users 6 Mar 24 05:37 iscolored
-rw-r--r-- 1 mcc users 211K Mar 24 05:37 model.pnml
--------------------
content from stdout:
=== Data for post analysis generated by BenchKit (invocation template)
The expected result is a vector of booleans
BOOL_VECTOR
here is the order used to build the result vector(from text file)
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-00
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-01
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-02
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-03
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-04
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-05
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-06
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-07
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-08
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-09
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-10
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-11
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-12
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-13
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-14
FORMULA_NAME DNAwalker-PT-16redondantChoiceR-15
=== Now, execution of the tool begins
BK_START 1589304523741
----------------------------------------------------------------------
GreatSPN-meddly tool, MCC 2020
----------------------------------------------------------------------
Running DNAwalker (PT), instance 16redondantChoiceR
LOADING PETRI NET FILE /home/mcc/execution/model.pnml (PNML) ...
PNML VERSION 2009, P/T NET.
COLOR CLASSES: 0
CONSTANTS: 0
PLACES: 43
TRANSITIONS: 490
COLOR VARS: 0
MEASURES: 0
LOADING TIME: [User 0.007s, Sys 0.000s]
SAVING FILE /home/mcc/execution/model (.net / .def) ...
EXPORT TIME: [User 0.001s, Sys 0.000s]
----------------------------------------------------------------------
GreatSPN/Meddly.
Copyright (C) 1987-2020, University of Torino, Italy.
Based on MEDDLY version 0.16.0
Copyright (C) 2009, Iowa State University Research Foundation, Inc.
website: http://meddly.sourceforge.net
Process ID: 456
MODEL NAME: /home/mcc/execution/model
43 places, 490 transitions.
FORMULA DNAwalker-PT-16redondantChoiceR-01 FALSE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
FORMULA DNAwalker-PT-16redondantChoiceR-06 TRUE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
FORMULA DNAwalker-PT-16redondantChoiceR-05 FALSE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
FORMULA DNAwalker-PT-16redondantChoiceR-10 TRUE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
FORMULA DNAwalker-PT-16redondantChoiceR-09 FALSE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
FORMULA DNAwalker-PT-16redondantChoiceR-14 FALSE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
FORMULA DNAwalker-PT-16redondantChoiceR-15 FALSE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
FORMULA DNAwalker-PT-16redondantChoiceR-03 TRUE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
FORMULA DNAwalker-PT-16redondantChoiceR-00 FALSE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
FORMULA DNAwalker-PT-16redondantChoiceR-02 FALSE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
FORMULA DNAwalker-PT-16redondantChoiceR-08 FALSE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
FORMULA DNAwalker-PT-16redondantChoiceR-07 FALSE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
FORMULA DNAwalker-PT-16redondantChoiceR-12 FALSE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
FORMULA DNAwalker-PT-16redondantChoiceR-11 FALSE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
FORMULA DNAwalker-PT-16redondantChoiceR-04 FALSE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
FORMULA DNAwalker-PT-16redondantChoiceR-13 FALSE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
Ok.
EXITCODE: 0
----------------------------------------------------------------------
BK_STOP 1589305580599
--------------------
content from stderr:
Sequence of Actions to be Executed by the VM
This is useful if one wants to reexecute the tool in the VM from the submitted image disk.
set -x
# this is for BenchKit: configuration of major elements for the test
export BK_INPUT="DNAwalker-PT-16redondantChoiceR"
export BK_EXAMINATION="LTLCardinality"
export BK_TOOL="gspn"
export BK_RESULT_DIR="/tmp/BK_RESULTS/OUTPUTS"
export BK_TIME_CONFINEMENT="3600"
export BK_MEMORY_CONFINEMENT="16384"
# this is specific to your benchmark or test
export BIN_DIR="$HOME/BenchKit/bin"
# remove the execution directoty if it exists (to avoid increse of .vmdk images)
if [ -d execution ] ; then
rm -rf execution
fi
# this is for BenchKit: explicit launching of the test
echo "====================================================================="
echo " Generated by BenchKit 2-4028"
echo " Executing tool gspn"
echo " Input is DNAwalker-PT-16redondantChoiceR, examination is LTLCardinality"
echo " Time confinement is $BK_TIME_CONFINEMENT seconds"
echo " Memory confinement is 16384 MBytes"
echo " Number of cores is 4"
echo " Run identifier is r065-tajo-158922812400201"
echo "====================================================================="
echo
echo "--------------------"
echo "preparation of the directory to be used:"
tar xzf /home/mcc/BenchKit/INPUTS/DNAwalker-PT-16redondantChoiceR.tgz
mv DNAwalker-PT-16redondantChoiceR execution
cd execution
if [ "LTLCardinality" = "ReachabilityDeadlock" ] || [ "LTLCardinality" = "UpperBounds" ] || [ "LTLCardinality" = "QuasiLiveness" ] || [ "LTLCardinality" = "StableMarking" ] || [ "LTLCardinality" = "Liveness" ] || [ "LTLCardinality" = "OneSafe" ] || [ "LTLCardinality" = "StateSpace" ]; then
rm -f GenericPropertiesVerdict.xml
fi
pwd
ls -lh
echo
echo "--------------------"
echo "content from stdout:"
echo
echo "=== Data for post analysis generated by BenchKit (invocation template)"
echo
if [ "LTLCardinality" = "UpperBounds" ] ; then
echo "The expected result is a vector of positive values"
echo NUM_VECTOR
elif [ "LTLCardinality" != "StateSpace" ] ; then
echo "The expected result is a vector of booleans"
echo BOOL_VECTOR
else
echo "no data necessary for post analysis"
fi
echo
if [ -f "LTLCardinality.txt" ] ; then
echo "here is the order used to build the result vector(from text file)"
for x in $(grep Property LTLCardinality.txt | cut -d ' ' -f 2 | sort -u) ; do
echo "FORMULA_NAME $x"
done
elif [ -f "LTLCardinality.xml" ] ; then # for cunf (txt files deleted;-)
echo echo "here is the order used to build the result vector(from xml file)"
for x in $(grep '
echo "FORMULA_NAME $x"
done
elif [ "LTLCardinality" = "ReachabilityDeadlock" ] || [ "LTLCardinality" = "QuasiLiveness" ] || [ "LTLCardinality" = "StableMarking" ] || [ "LTLCardinality" = "Liveness" ] || [ "LTLCardinality" = "OneSafe" ] ; then
echo "FORMULA_NAME LTLCardinality"
fi
echo
echo "=== Now, execution of the tool begins"
echo
echo -n "BK_START "
date -u +%s%3N
echo
timeout -s 9 $BK_TIME_CONFINEMENT bash -c "/home/mcc/BenchKit/BenchKit_head.sh 2> STDERR ; echo ; echo -n \"BK_STOP \" ; date -u +%s%3N"
if [ $? -eq 137 ] ; then
echo
echo "BK_TIME_CONFINEMENT_REACHED"
fi
echo
echo "--------------------"
echo "content from stderr:"
echo
cat STDERR ;

