About the Execution of Irma.struct for DNAwalker-PT-13ringRLLarge
| Execution Summary | |||||
| Max Memory Used (MB) |
Time wait (ms) | CPU Usage (ms) | I/O Wait (ms) | Computed Result | Execution Status |
| 10115.430 | 126910.00 | 126586.00 | 993.10 | TFFFTTTTTFFFTFTF | normal |
Execution Chart
We display below the execution chart for this examination (boot time has been removed).
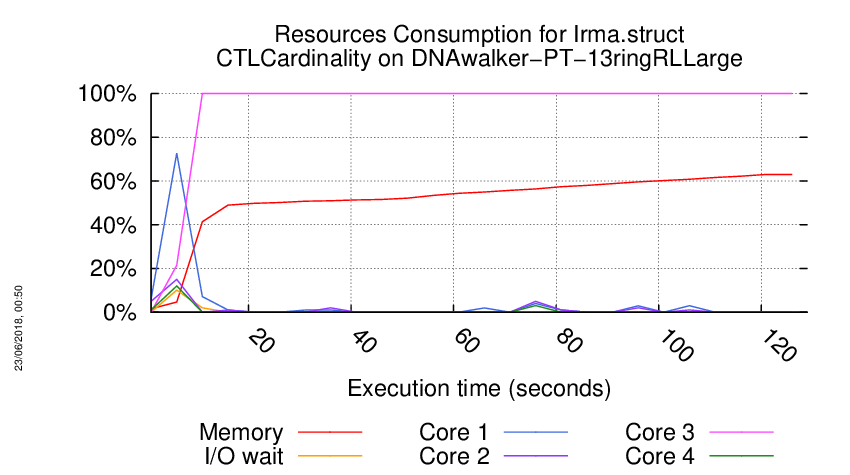
Trace from the execution
Waiting for the VM to be ready (probing ssh)
.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................
/home/mcc/execution
total 304K
-rw-r--r-- 1 mcc users 3.2K May 15 18:54 CTLCardinality.txt
-rw-r--r-- 1 mcc users 18K May 15 18:54 CTLCardinality.xml
-rw-r--r-- 1 mcc users 2.6K May 15 18:54 CTLFireability.txt
-rw-r--r-- 1 mcc users 15K May 15 18:54 CTLFireability.xml
-rw-r--r-- 1 mcc users 4.0K May 15 18:50 GenericPropertiesDefinition.xml
-rw-r--r-- 1 mcc users 5.6K May 15 18:50 GenericPropertiesVerdict.xml
-rw-r--r-- 1 mcc users 2.4K May 15 18:54 LTLCardinality.txt
-rw-r--r-- 1 mcc users 11K May 15 18:54 LTLCardinality.xml
-rw-r--r-- 1 mcc users 2.0K May 15 18:54 LTLFireability.txt
-rw-r--r-- 1 mcc users 9.0K May 15 18:54 LTLFireability.xml
-rw-r--r-- 1 mcc users 3.9K May 15 18:54 ReachabilityCardinality.txt
-rw-r--r-- 1 mcc users 22K May 15 18:54 ReachabilityCardinality.xml
-rw-r--r-- 1 mcc users 117 May 15 18:54 ReachabilityDeadlock.txt
-rw-r--r-- 1 mcc users 355 May 15 18:54 ReachabilityDeadlock.xml
-rw-r--r-- 1 mcc users 3.1K May 15 18:54 ReachabilityFireability.txt
-rw-r--r-- 1 mcc users 19K May 15 18:54 ReachabilityFireability.xml
-rw-r--r-- 1 mcc users 1.7K May 15 18:54 UpperBounds.txt
-rw-r--r-- 1 mcc users 3.8K May 15 18:54 UpperBounds.xml
-rw-r--r-- 1 mcc users 6 May 15 18:50 equiv_col
-rw-r--r-- 1 mcc users 14 May 15 18:50 instance
-rw-r--r-- 1 mcc users 6 May 15 18:50 iscolored
-rw-r--r-- 1 mcc users 135K May 15 18:50 model.pnml
=====================================================================
Generated by BenchKit 2-3637
Executing tool irma4mcc-structural
Input is DNAwalker-PT-13ringRLLarge, examination is CTLCardinality
Time confinement is 3600 seconds
Memory confinement is 16384 MBytes
Number of cores is 4
Run identifier is r067-smll-152649739900311
=====================================================================
--------------------
content from stdout:
=== Data for post analysis generated by BenchKit (invocation template)
The expected result is a vector of booleans
BOOL_VECTOR
here is the order used to build the result vector(from text file)
FORMULA_NAME DNAwalker-PT-13ringRLLarge-CTLCardinality-00
FORMULA_NAME DNAwalker-PT-13ringRLLarge-CTLCardinality-01
FORMULA_NAME DNAwalker-PT-13ringRLLarge-CTLCardinality-02
FORMULA_NAME DNAwalker-PT-13ringRLLarge-CTLCardinality-03
FORMULA_NAME DNAwalker-PT-13ringRLLarge-CTLCardinality-04
FORMULA_NAME DNAwalker-PT-13ringRLLarge-CTLCardinality-05
FORMULA_NAME DNAwalker-PT-13ringRLLarge-CTLCardinality-06
FORMULA_NAME DNAwalker-PT-13ringRLLarge-CTLCardinality-07
FORMULA_NAME DNAwalker-PT-13ringRLLarge-CTLCardinality-08
FORMULA_NAME DNAwalker-PT-13ringRLLarge-CTLCardinality-09
FORMULA_NAME DNAwalker-PT-13ringRLLarge-CTLCardinality-10
FORMULA_NAME DNAwalker-PT-13ringRLLarge-CTLCardinality-11
FORMULA_NAME DNAwalker-PT-13ringRLLarge-CTLCardinality-12
FORMULA_NAME DNAwalker-PT-13ringRLLarge-CTLCardinality-13
FORMULA_NAME DNAwalker-PT-13ringRLLarge-CTLCardinality-14
FORMULA_NAME DNAwalker-PT-13ringRLLarge-CTLCardinality-15
=== Now, execution of the tool begins
BK_START 1526663895705
BK_STOP 1526664022615
--------------------
content from stderr:
Prefix is 75f5f979.
Reading known information in /usr/share/mcc4mcc/75f5f979-known.json.
Reading learned information in /usr/share/mcc4mcc/75f5f979-learned.json.
Reading value translations in /usr/share/mcc4mcc/75f5f979-values.json.
Using directory /home/mcc/execution for input, as it contains a model.pnml file.
Using DNAwalker-PT-13ringRLLarge as instance name.
Using DNAwalker as model name.
Using algorithm or tool bmdt.
Model characteristics are: {'Examination': 'CTLCardinality', 'Place/Transition': True, 'Colored': False, 'Relative-Time': 1, 'Relative-Memory': 1, 'Ordinary': False, 'Simple Free Choice': False, 'Extended Free Choice': False, 'State Machine': False, 'Marked Graph': False, 'Connected': True, 'Strongly Connected': False, 'Source Place': None, 'Sink Place': False, 'Source Transition': False, 'Sink Transition': None, 'Loop Free': False, 'Conservative': False, 'Sub-Conservative': True, 'Nested Units': False, 'Safe': False, 'Deadlock': True, 'Reversible': False, 'Quasi Live': None, 'Live': None}.
Known tools are: [{'Time': 138932, 'Memory': 9671.25, 'Tool': 'marcie'}, {'Time': 143622, 'Memory': 9671.41, 'Tool': 'marcie'}, {'Time': 306618, 'Memory': 2846.91, 'Tool': 'itstools'}, {'Time': 314640, 'Memory': 2857.11, 'Tool': 'itstools'}, {'Time': 1332653, 'Memory': 7152.3, 'Tool': 'lola'}, {'Time': 1332740, 'Memory': 7175.24, 'Tool': 'lola'}].
Learned tools are: [{'Tool': 'lola'}].
Learned tool lola is 9.592124204646877x far from the best tool marcie.
CTLCardinality marcie DNAwalker-PT-13ringRLLarge...
timeout --kill-after=10s --signal=SIGINT 1m for testing only
Marcie rev. 8852M (built: crohr on 2017-05-03)
A model checker for Generalized Stochastic Petri nets
authors: Alex Tovchigrechko (IDD package and CTL model checking)
Martin Schwarick (Symbolic numerical analysis and CSL model checking)
Christian Rohr (Simulative and approximative numerical model checking)
marcie@informatik.tu-cottbus.de
called as: marcie --net-file=model.pnml --mcc-file=CTLCardinality.xml --memory=6
parse successfull
net created successfully
Net: DNAwalker_PT_13ringRLLarge
(NrP: 33 NrTr: 312 NrArc: 916)
check for maximal unmarked siphon
ok
check for constant places
ok
parse formulas
formulas created successfully
place and transition orderings generation:0m 0.001sec
check if there are places and transitions
ok
check if there are transitions without pre-places
ok
check if at least one transition is enabled in m0
ok
check if there are transitions that can never fire
ok
net check time: 0m 0.000sec
init dd package: 0m 1.745sec
ptnet_zbdd.cc:255: Boundedness exception: net is not 1-bounded!
parse successfull
net created successfully
Net: DNAwalker_PT_13ringRLLarge
(NrP: 33 NrTr: 312 NrArc: 916)
check for maximal unmarked siphon
ok
check for constant places
ok
check if there are places and transitions
ok
check if there are transitions without pre-places
ok
check if at least one transition is enabled in m0
ok
check if there are transitions that can never fire
parse formulas
formulas created successfully
place and transition orderings generation:0m 0.000sec
ok
net check time: 0m 0.000sec
init dd package: 0m 4.983sec
RS generation: 0m32.783sec
-> reachability set: #nodes 15938 (1.6e+04) #states 1,885,372,776 (9)
starting MCC model checker
--------------------------
checking: EX [EG [~ [1<=B16]]]
normalized: EX [EG [~ [1<=B16]]]
abstracting: (1<=B16)
states: 556,663,898 (8)
.
EG iterations: 1
.-> the formula is TRUE
FORMULA DNAwalker-PT-13ringRLLarge-CTLCardinality-06 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 4.509sec
checking: ~ [AG [AF [2<=A11]]]
normalized: E [true U EG [~ [2<=A11]]]
abstracting: (2<=A11)
states: 73,673,326 (7)
.
EG iterations: 1
-> the formula is TRUE
FORMULA DNAwalker-PT-13ringRLLarge-CTLCardinality-07 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.188sec
checking: EX [AF [3<=A22]]
normalized: EX [~ [EG [~ [3<=A22]]]]
abstracting: (3<=A22)
states: 0
EG iterations: 0
.-> the formula is FALSE
FORMULA DNAwalker-PT-13ringRLLarge-CTLCardinality-11 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.012sec
checking: ~ [AF [~ [~ [2<=A5]]]]
normalized: EG [~ [2<=A5]]
abstracting: (2<=A5)
states: 47,796,541 (7)
.
EG iterations: 1
-> the formula is TRUE
FORMULA DNAwalker-PT-13ringRLLarge-CTLCardinality-14 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.055sec
checking: A [EX [1<=A2] U AX [2<=A17]]
normalized: [~ [E [EX [~ [2<=A17]] U [~ [EX [1<=A2]] & EX [~ [2<=A17]]]]] & ~ [EG [EX [~ [2<=A17]]]]]
abstracting: (2<=A17)
states: 48,083,646 (7)
.
EG iterations: 0
abstracting: (2<=A17)
states: 48,083,646 (7)
.abstracting: (1<=A2)
states: 861,987,750 (8)
.abstracting: (2<=A17)
states: 48,083,646 (7)
.-> the formula is FALSE
FORMULA DNAwalker-PT-13ringRLLarge-CTLCardinality-13 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 7.657sec
checking: [AF [AX [3<=A4]] & EF [AF [B18<=A14]]]
normalized: [~ [EG [EX [~ [3<=A4]]]] & E [true U ~ [EG [~ [B18<=A14]]]]]
abstracting: (B18<=A14)
states: 1,574,706,224 (9)
.
EG iterations: 1
abstracting: (3<=A4)
states: 0
.
EG iterations: 0
-> the formula is FALSE
FORMULA DNAwalker-PT-13ringRLLarge-CTLCardinality-01 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 2.199sec
checking: AF [AF [[A17<=B16 & 1<=A17]]]
normalized: ~ [EG [EG [~ [[A17<=B16 & 1<=A17]]]]]
abstracting: (1<=A17)
states: 632,196,820 (8)
abstracting: (A17<=B16)
states: 1,421,330,379 (9)
.
EG iterations: 1
.
EG iterations: 1
-> the formula is FALSE
FORMULA DNAwalker-PT-13ringRLLarge-CTLCardinality-02 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 1.714sec
checking: ~ [~ [EG [[A18<=A12 | 1<=A6]]]]
normalized: EG [[A18<=A12 | 1<=A6]]
abstracting: (1<=A6)
states: 953,311,836 (8)
abstracting: (A18<=A12)
states: 1,542,921,990 (9)
.......
EG iterations: 7
-> the formula is TRUE
FORMULA DNAwalker-PT-13ringRLLarge-CTLCardinality-04 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 2.838sec
checking: [EG [AF [A23<=A11]] & AF [AG [3<=A11]]]
normalized: [~ [EG [E [true U ~ [3<=A11]]]] & EG [~ [EG [~ [A23<=A11]]]]]
abstracting: (A23<=A11)
states: 1,424,603,283 (9)
........
EG iterations: 8
........
EG iterations: 8
abstracting: (3<=A11)
states: 0
EG iterations: 0
-> the formula is FALSE
FORMULA DNAwalker-PT-13ringRLLarge-CTLCardinality-10 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 9.222sec
checking: AX [[AG [1<=A24] | [~ [A23<=B16] | ~ [1<=A4]]]]
normalized: ~ [EX [~ [[[~ [1<=A4] | ~ [A23<=B16]] | ~ [E [true U ~ [1<=A24]]]]]]]
abstracting: (1<=A24)
states: 976,132,633 (8)
abstracting: (A23<=B16)
states: 1,233,470,044 (9)
abstracting: (1<=A4)
states: 642,168,245 (8)
.-> the formula is FALSE
FORMULA DNAwalker-PT-13ringRLLarge-CTLCardinality-03 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m20.927sec
checking: EG [[AF [A22<=A18] | EX [A23<=A2]]]
normalized: EG [[EX [A23<=A2] | ~ [EG [~ [A22<=A18]]]]]
abstracting: (A22<=A18)
states: 592,707,084 (8)
.
EG iterations: 1
abstracting: (A23<=A2)
states: 1,354,510,905 (9)
........
EG iterations: 7
-> the formula is TRUE
FORMULA DNAwalker-PT-13ringRLLarge-CTLCardinality-05 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 6.895sec
checking: [EG [~ [[3<=A23 & A19<=A18]]] | ~ [~ [B4<=A17]]]
normalized: [B4<=A17 | EG [~ [[3<=A23 & A19<=A18]]]]
abstracting: (A19<=A18)
states: 1,227,500,089 (9)
abstracting: (3<=A23)
states: 0
EG iterations: 0
abstracting: (B4<=A17)
states: 1,490,503,274 (9)
-> the formula is TRUE
FORMULA DNAwalker-PT-13ringRLLarge-CTLCardinality-08 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.072sec
checking: EF [[EG [1<=B18] & [[A27<=B5 | 2<=A22] | A27<=A16]]]
normalized: E [true U [EG [1<=B18] & [A27<=A16 | [A27<=B5 | 2<=A22]]]]
abstracting: (2<=A22)
states: 152,467,108 (8)
abstracting: (A27<=B5)
states: 1,224,650,570 (9)
abstracting: (A27<=A16)
states: 1,230,010,070 (9)
abstracting: (1<=B18)
states: 594,446,472 (8)
.
EG iterations: 1
-> the formula is TRUE
FORMULA DNAwalker-PT-13ringRLLarge-CTLCardinality-00 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 6.535sec
checking: [[2<=A2 | A17<=A12] & AF [~ [[A10<=A13 | 1<=A10]]]]
normalized: [~ [EG [[A10<=A13 | 1<=A10]]] & [2<=A2 | A17<=A12]]
abstracting: (A17<=A12)
states: 1,551,009,346 (9)
abstracting: (2<=A2)
states: 69,365,904 (7)
abstracting: (1<=A10)
states: 882,354,750 (8)
abstracting: (A10<=A13)
states: 1,415,876,459 (9)
EG iterations: 0
-> the formula is FALSE
FORMULA DNAwalker-PT-13ringRLLarge-CTLCardinality-15 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.032sec
checking: [E [[A20<=B17 | 1<=A7] U [1<=A1 & 2<=A2]] | ~ [E [1<=A3 U 2<=A5]]]
normalized: [~ [E [1<=A3 U 2<=A5]] | E [[A20<=B17 | 1<=A7] U [1<=A1 & 2<=A2]]]
abstracting: (2<=A2)
states: 69,365,904 (7)
abstracting: (1<=A1)
states: 729
abstracting: (1<=A7)
states: 973,214,356 (8)
abstracting: (A20<=B17)
states: 1,185,288,227 (9)
abstracting: (2<=A5)
states: 47,796,541 (7)
abstracting: (1<=A3)
states: 907,623,618 (8)
-> the formula is FALSE
FORMULA DNAwalker-PT-13ringRLLarge-CTLCardinality-09 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 4.094sec
checking: [~ [AF [[A24<=A5 | 2<=A21]]] & E [[3<=B16 | 2<=A7] U A4<=A19]]
normalized: [E [[3<=B16 | 2<=A7] U A4<=A19] & EG [~ [[A24<=A5 | 2<=A21]]]]
abstracting: (2<=A21)
states: 70,395,378 (7)
abstracting: (A24<=A5)
states: 1,220,020,436 (9)
........
EG iterations: 8
abstracting: (A4<=A19)
states: 1,544,716,414 (9)
abstracting: (2<=A7)
states: 70,675,984 (7)
abstracting: (3<=B16)
states: 0
-> the formula is TRUE
FORMULA DNAwalker-PT-13ringRLLarge-CTLCardinality-12 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 9.366sec
totally nodes used: 21259832 (2.1e+07)
number of garbage collections: 0
fire ops cache: hits/miss/sum: 81216837 99908552 181125389
used/not used/entry size/cache size: 56444010 10664854 16 1024MB
basic ops cache: hits/miss/sum: 13913692 13026710 26940402
used/not used/entry size/cache size: 12263933 4513283 12 192MB
unary ops cache: hits/miss/sum: 0 0 0
used/not used/entry size/cache size: 0 16777216 8 128MB
abstract ops cache: hits/miss/sum: 0 0 0
used/not used/entry size/cache size: 0 16777216 12 192MB
state nr cache: hits/miss/sum: 192526 205039 397565
used/not used/entry size/cache size: 202589 8186019 32 256MB
max state cache: hits/miss/sum: 0 0 0
used/not used/entry size/cache size: 0 8388608 32 256MB
uniqueHash elements/entry size/size: 67108864 4 256MB
0 49919282
1 13973289
2 2630496
3 438777
4 91444
5 27099
6 16174
7 5358
8 2821
9 1230
>= 10 2894
Total processing time: 2m 1.236sec
.........10 71817.........20 44331............................10 23677....................
Sequence of Actions to be Executed by the VM
This is useful if one wants to reexecute the tool in the VM from the submitted image disk.
set -x
# this is for BenchKit: configuration of major elements for the test
export BK_INPUT="DNAwalker-PT-13ringRLLarge"
export BK_EXAMINATION="CTLCardinality"
export BK_TOOL="irma4mcc-structural"
export BK_RESULT_DIR="/tmp/BK_RESULTS/OUTPUTS"
export BK_TIME_CONFINEMENT="3600"
export BK_MEMORY_CONFINEMENT="16384"
# this is specific to your benchmark or test
export BIN_DIR="$HOME/BenchKit/bin"
# remove the execution directoty if it exists (to avoid increse of .vmdk images)
if [ -d execution ] ; then
rm -rf execution
fi
tar xzf /home/mcc/BenchKit/INPUTS/DNAwalker-PT-13ringRLLarge.tgz
mv DNAwalker-PT-13ringRLLarge execution
cd execution
pwd
ls -lh
# this is for BenchKit: explicit launching of the test
echo "====================================================================="
echo " Generated by BenchKit 2-3637"
echo " Executing tool irma4mcc-structural"
echo " Input is DNAwalker-PT-13ringRLLarge, examination is CTLCardinality"
echo " Time confinement is $BK_TIME_CONFINEMENT seconds"
echo " Memory confinement is 16384 MBytes"
echo " Number of cores is 4"
echo " Run identifier is r067-smll-152649739900311"
echo "====================================================================="
echo
echo "--------------------"
echo "content from stdout:"
echo
echo "=== Data for post analysis generated by BenchKit (invocation template)"
echo
if [ "CTLCardinality" = "UpperBounds" ] ; then
echo "The expected result is a vector of positive values"
echo NUM_VECTOR
elif [ "CTLCardinality" != "StateSpace" ] ; then
echo "The expected result is a vector of booleans"
echo BOOL_VECTOR
else
echo "no data necessary for post analysis"
fi
echo
if [ -f "CTLCardinality.txt" ] ; then
echo "here is the order used to build the result vector(from text file)"
for x in $(grep Property CTLCardinality.txt | cut -d ' ' -f 2 | sort -u) ; do
echo "FORMULA_NAME $x"
done
elif [ -f "CTLCardinality.xml" ] ; then # for cunf (txt files deleted;-)
echo echo "here is the order used to build the result vector(from xml file)"
for x in $(grep '
echo "FORMULA_NAME $x"
done
fi
echo
echo "=== Now, execution of the tool begins"
echo
echo -n "BK_START "
date -u +%s%3N
echo
timeout -s 9 $BK_TIME_CONFINEMENT bash -c "/home/mcc/BenchKit/BenchKit_head.sh 2> STDERR ; echo ; echo -n \"BK_STOP \" ; date -u +%s%3N"
if [ $? -eq 137 ] ; then
echo
echo "BK_TIME_CONFINEMENT_REACHED"
fi
echo
echo "--------------------"
echo "content from stderr:"
echo
cat STDERR ;

