About the Execution of Irma.full for DNAwalker-PT-09ringLR
| Execution Summary | |||||
| Max Memory Used (MB) |
Time wait (ms) | CPU Usage (ms) | I/O Wait (ms) | Computed Result | Execution Status |
| 499.800 | 10126.00 | 10162.00 | 935.40 | 2.8210E+0007 2.4089E+0008 2 22 | normal |
Execution Chart
We display below the execution chart for this examination (boot time has been removed).
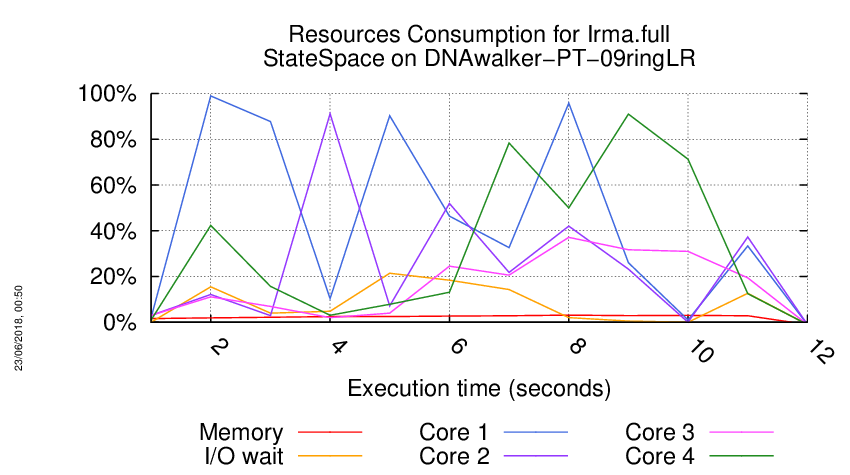
Trace from the execution
Waiting for the VM to be ready (probing ssh)
.......................................................................................................................................................................................................................................................................................................................................................................................................
/home/mcc/execution
total 276K
-rw-r--r-- 1 mcc users 3.0K May 15 18:54 CTLCardinality.txt
-rw-r--r-- 1 mcc users 16K May 15 18:54 CTLCardinality.xml
-rw-r--r-- 1 mcc users 2.5K May 15 18:54 CTLFireability.txt
-rw-r--r-- 1 mcc users 16K May 15 18:54 CTLFireability.xml
-rw-r--r-- 1 mcc users 4.0K May 15 18:50 GenericPropertiesDefinition.xml
-rw-r--r-- 1 mcc users 5.6K May 15 18:50 GenericPropertiesVerdict.xml
-rw-r--r-- 1 mcc users 2.4K May 15 18:54 LTLCardinality.txt
-rw-r--r-- 1 mcc users 9.9K May 15 18:54 LTLCardinality.xml
-rw-r--r-- 1 mcc users 1.9K May 15 18:54 LTLFireability.txt
-rw-r--r-- 1 mcc users 8.3K May 15 18:54 LTLFireability.xml
-rw-r--r-- 1 mcc users 4.0K May 15 18:54 ReachabilityCardinality.txt
-rw-r--r-- 1 mcc users 22K May 15 18:54 ReachabilityCardinality.xml
-rw-r--r-- 1 mcc users 112 May 15 18:54 ReachabilityDeadlock.txt
-rw-r--r-- 1 mcc users 350 May 15 18:54 ReachabilityDeadlock.xml
-rw-r--r-- 1 mcc users 2.9K May 15 18:54 ReachabilityFireability.txt
-rw-r--r-- 1 mcc users 17K May 15 18:54 ReachabilityFireability.xml
-rw-r--r-- 1 mcc users 1.7K May 15 18:54 UpperBounds.txt
-rw-r--r-- 1 mcc users 3.7K May 15 18:54 UpperBounds.xml
-rw-r--r-- 1 mcc users 6 May 15 18:50 equiv_col
-rw-r--r-- 1 mcc users 9 May 15 18:50 instance
-rw-r--r-- 1 mcc users 6 May 15 18:50 iscolored
-rw-r--r-- 1 mcc users 112K May 15 18:50 model.pnml
=====================================================================
Generated by BenchKit 2-3637
Executing tool irma4mcc-full
Input is DNAwalker-PT-09ringLR, examination is StateSpace
Time confinement is 3600 seconds
Memory confinement is 16384 MBytes
Number of cores is 4
Run identifier is r066-smll-152649737300281
=====================================================================
--------------------
content from stdout:
=== Data for post analysis generated by BenchKit (invocation template)
no data necessary for post analysis
=== Now, execution of the tool begins
BK_START 1526664483967
BK_STOP 1526664494093
--------------------
content from stderr:
Prefix is 65b80f64.
Reading known information in /usr/share/mcc4mcc/65b80f64-known.json.
Reading learned information in /usr/share/mcc4mcc/65b80f64-learned.json.
Reading value translations in /usr/share/mcc4mcc/65b80f64-values.json.
Using directory /home/mcc/execution for input, as it contains a model.pnml file.
Using DNAwalker-PT-09ringLR as instance name.
Using DNAwalker as model name.
Using algorithm or tool bmdt.
Model characteristics are: {'Examination': 'StateSpace', 'Place/Transition': True, 'Colored': False, 'Relative-Time': 1, 'Relative-Memory': 1, 'Ordinary': False, 'Simple Free Choice': False, 'Extended Free Choice': False, 'State Machine': False, 'Marked Graph': False, 'Connected': True, 'Strongly Connected': False, 'Source Place': None, 'Sink Place': False, 'Source Transition': False, 'Sink Transition': None, 'Loop Free': False, 'Conservative': False, 'Sub-Conservative': True, 'Nested Units': False, 'Safe': False, 'Deadlock': True, 'Reversible': False, 'Quasi Live': None, 'Live': None}.
Known tools are: [{'Time': 1239, 'Memory': 134.22, 'Tool': 'gspn'}, {'Time': 2236, 'Memory': 1875.07, 'Tool': 'tina'}, {'Time': 2616, 'Memory': 277.1, 'Tool': 'itstools'}, {'Time': 2617, 'Memory': 235.48, 'Tool': 'itstools'}, {'Time': 3173, 'Memory': 2604.71, 'Tool': 'tina'}, {'Time': 3874, 'Memory': 175.86, 'Tool': 'gspn'}, {'Time': 8865, 'Memory': 7515.51, 'Tool': 'marcie'}, {'Time': 9412, 'Memory': 7515.66, 'Tool': 'marcie'}, {'Time': 493516, 'Memory': 15499.28, 'Tool': 'tina'}, {'Time': 515364, 'Memory': 15499.17, 'Tool': 'tina'}].
Learned tools are: [{'Tool': 'gspn'}].
Learned tool gspn is 1.0x far from the best tool gspn.
StateSpace gspn DNAwalker-PT-09ringLR...
LOADING model.pnml ...
MODEL CLASS: P/T NET
PLACES: 27
TRANSITIONS: 260
CONSTANTS: 0
TEMPLATE VARS: 0
ARCS: 760
LOADING TIME: 0.592
SAVING AS model.(net/def) ...
SAVING TIME: 0.069
SAVING NAME MAP FILE model.id2name ...
TOTAL TIME: 1.388
OK.
Can't open file model.pin for r
GreatSPN/Meddly.
Copyright (C) 1987-2017, University of Torino, Italy.
Based on MEDDLY version 0.14.765
Copyright (C) 2009, Iowa State University Research Foundation, Inc.
website: http://meddly.sourceforge.net
Process ID: 29
MODEL NAME: model
27 places, 260 transitions.
Setting MEDDLY cache to 67108864 entries.
STATE_SPACE STATES 28209796 TECHNIQUES DECISION_DIAGRAMS SEQUENTIAL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
STATE_SPACE MAX_TOKEN_PER_MARKING 22 TECHNIQUES DECISION_DIAGRAMS SEQUENTIAL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
STATE_SPACE MAX_TOKEN_IN_PLACE 2 TECHNIQUES DECISION_DIAGRAMS SEQUENTIAL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
STATE_SPACE TRANSITIONS 240889487 TECHNIQUES DECISION_DIAGRAMS SEQUENTIAL_PROCESSING UNFOLDING_TO_PT USE_NUPN TOPOLOGICAL
Sequence of Actions to be Executed by the VM
This is useful if one wants to reexecute the tool in the VM from the submitted image disk.
set -x
# this is for BenchKit: configuration of major elements for the test
export BK_INPUT="DNAwalker-PT-09ringLR"
export BK_EXAMINATION="StateSpace"
export BK_TOOL="irma4mcc-full"
export BK_RESULT_DIR="/tmp/BK_RESULTS/OUTPUTS"
export BK_TIME_CONFINEMENT="3600"
export BK_MEMORY_CONFINEMENT="16384"
# this is specific to your benchmark or test
export BIN_DIR="$HOME/BenchKit/bin"
# remove the execution directoty if it exists (to avoid increse of .vmdk images)
if [ -d execution ] ; then
rm -rf execution
fi
tar xzf /home/mcc/BenchKit/INPUTS/DNAwalker-PT-09ringLR.tgz
mv DNAwalker-PT-09ringLR execution
cd execution
pwd
ls -lh
# this is for BenchKit: explicit launching of the test
echo "====================================================================="
echo " Generated by BenchKit 2-3637"
echo " Executing tool irma4mcc-full"
echo " Input is DNAwalker-PT-09ringLR, examination is StateSpace"
echo " Time confinement is $BK_TIME_CONFINEMENT seconds"
echo " Memory confinement is 16384 MBytes"
echo " Number of cores is 4"
echo " Run identifier is r066-smll-152649737300281"
echo "====================================================================="
echo
echo "--------------------"
echo "content from stdout:"
echo
echo "=== Data for post analysis generated by BenchKit (invocation template)"
echo
if [ "StateSpace" = "UpperBounds" ] ; then
echo "The expected result is a vector of positive values"
echo NUM_VECTOR
elif [ "StateSpace" != "StateSpace" ] ; then
echo "The expected result is a vector of booleans"
echo BOOL_VECTOR
else
echo "no data necessary for post analysis"
fi
echo
if [ -f "StateSpace.txt" ] ; then
echo "here is the order used to build the result vector(from text file)"
for x in $(grep Property StateSpace.txt | cut -d ' ' -f 2 | sort -u) ; do
echo "FORMULA_NAME $x"
done
elif [ -f "StateSpace.xml" ] ; then # for cunf (txt files deleted;-)
echo echo "here is the order used to build the result vector(from xml file)"
for x in $(grep '
echo "FORMULA_NAME $x"
done
fi
echo
echo "=== Now, execution of the tool begins"
echo
echo -n "BK_START "
date -u +%s%3N
echo
timeout -s 9 $BK_TIME_CONFINEMENT bash -c "/home/mcc/BenchKit/BenchKit_head.sh 2> STDERR ; echo ; echo -n \"BK_STOP \" ; date -u +%s%3N"
if [ $? -eq 137 ] ; then
echo
echo "BK_TIME_CONFINEMENT_REACHED"
fi
echo
echo "--------------------"
echo "content from stderr:"
echo
cat STDERR ;

