About the Execution of ITS-Tools.L for Angiogenesis-PT-05
| Execution Summary | |||||
| Max Memory Used (MB) |
Time wait (ms) | CPU Usage (ms) | I/O Wait (ms) | Computed Result | Execution Status |
| 15778.570 | 370990.00 | 368518.00 | 3694.60 | TFFFTTTFTTFFFFTF | normal |
Execution Chart
We display below the execution chart for this examination (boot time has been removed).
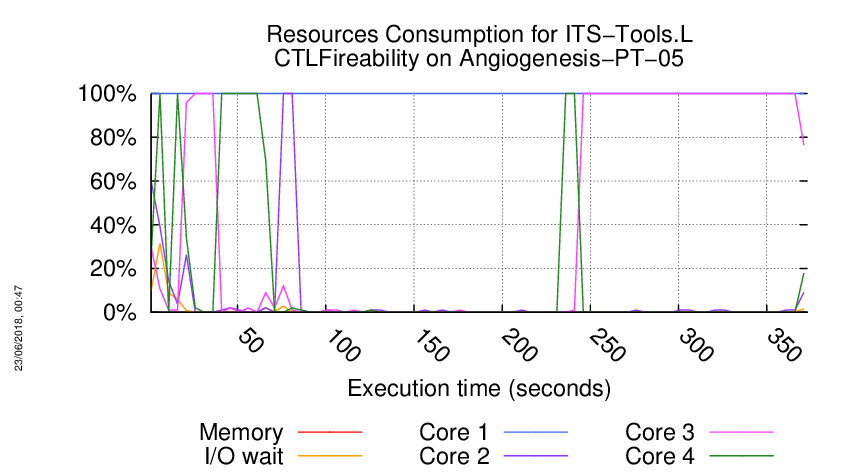
Trace from the execution
Waiting for the VM to be ready (probing ssh)
..............................................................................................
/home/mcc/execution
total 204K
-rw-r--r-- 1 mcc users 3.1K May 15 18:54 CTLCardinality.txt
-rw-r--r-- 1 mcc users 15K May 15 18:54 CTLCardinality.xml
-rw-r--r-- 1 mcc users 2.6K May 15 18:54 CTLFireability.txt
-rw-r--r-- 1 mcc users 19K May 15 18:54 CTLFireability.xml
-rw-r--r-- 1 mcc users 4.0K May 15 18:49 GenericPropertiesDefinition.xml
-rw-r--r-- 1 mcc users 6.0K May 15 18:49 GenericPropertiesVerdict.xml
-rw-r--r-- 1 mcc users 2.5K May 15 18:54 LTLCardinality.txt
-rw-r--r-- 1 mcc users 11K May 15 18:54 LTLCardinality.xml
-rw-r--r-- 1 mcc users 2.0K May 15 18:54 LTLFireability.txt
-rw-r--r-- 1 mcc users 11K May 15 18:54 LTLFireability.xml
-rw-r--r-- 1 mcc users 4.4K May 15 18:54 ReachabilityCardinality.txt
-rw-r--r-- 1 mcc users 21K May 15 18:54 ReachabilityCardinality.xml
-rw-r--r-- 1 mcc users 109 May 15 18:54 ReachabilityDeadlock.txt
-rw-r--r-- 1 mcc users 347 May 15 18:54 ReachabilityDeadlock.xml
-rw-r--r-- 1 mcc users 2.4K May 15 18:54 ReachabilityFireability.txt
-rw-r--r-- 1 mcc users 14K May 15 18:54 ReachabilityFireability.xml
-rw-r--r-- 1 mcc users 1.7K May 15 18:54 UpperBounds.txt
-rw-r--r-- 1 mcc users 3.8K May 15 18:54 UpperBounds.xml
-rw-r--r-- 1 mcc users 6 May 15 18:49 equiv_col
-rw-r--r-- 1 mcc users 3 May 15 18:49 instance
-rw-r--r-- 1 mcc users 6 May 15 18:49 iscolored
-rw-r--r-- 1 mcc users 33K May 15 18:49 model.pnml
=====================================================================
Generated by BenchKit 2-3637
Executing tool itstoolsl
Input is Angiogenesis-PT-05, examination is CTLFireability
Time confinement is 3600 seconds
Memory confinement is 16384 MBytes
Number of cores is 4
Run identifier is r021-qhx1-152646246200011
=====================================================================
--------------------
content from stdout:
=== Data for post analysis generated by BenchKit (invocation template)
The expected result is a vector of booleans
BOOL_VECTOR
here is the order used to build the result vector(from text file)
FORMULA_NAME Angiogenesis-PT-05-CTLFireability-00
FORMULA_NAME Angiogenesis-PT-05-CTLFireability-01
FORMULA_NAME Angiogenesis-PT-05-CTLFireability-02
FORMULA_NAME Angiogenesis-PT-05-CTLFireability-03
FORMULA_NAME Angiogenesis-PT-05-CTLFireability-04
FORMULA_NAME Angiogenesis-PT-05-CTLFireability-05
FORMULA_NAME Angiogenesis-PT-05-CTLFireability-06
FORMULA_NAME Angiogenesis-PT-05-CTLFireability-07
FORMULA_NAME Angiogenesis-PT-05-CTLFireability-08
FORMULA_NAME Angiogenesis-PT-05-CTLFireability-09
FORMULA_NAME Angiogenesis-PT-05-CTLFireability-10
FORMULA_NAME Angiogenesis-PT-05-CTLFireability-11
FORMULA_NAME Angiogenesis-PT-05-CTLFireability-12
FORMULA_NAME Angiogenesis-PT-05-CTLFireability-13
FORMULA_NAME Angiogenesis-PT-05-CTLFireability-14
FORMULA_NAME Angiogenesis-PT-05-CTLFireability-15
=== Now, execution of the tool begins
BK_START 1527200697704
Converted graph to binary with : CommandLine [args=[/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.louvain.binaries_1.0.0.201805241334/bin/convert-linux64, -i, /tmp/graph6250556946777253892.txt, -o, /tmp/graph6250556946777253892.bin, -w, /tmp/graph6250556946777253892.weights], workingDir=null]
Built communities with : CommandLine [args=[/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.louvain.binaries_1.0.0.201805241334/bin/louvain-linux64, /tmp/graph6250556946777253892.bin, -l, -1, -v, -w, /tmp/graph6250556946777253892.weights, -q, 0, -e, 0.001], workingDir=null]
Invoking ITS tools like this :CommandLine [args=[/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.itstools.binaries_1.0.0.201805241334/bin/its-ctl-linux64, --gc-threshold, 2000000, --quiet, -i, /home/mcc/execution/CTLFireability.pnml.gal, -t, CGAL, -ctl, /home/mcc/execution/CTLFireability.ctl], workingDir=/home/mcc/execution]
its-ctl command run as :
/home/mcc/BenchKit/itstools/plugins/fr.lip6.move.gal.itstools.binaries_1.0.0.201805241334/bin/its-ctl-linux64 --gc-threshold 2000000 --quiet -i /home/mcc/execution/CTLFireability.pnml.gal -t CGAL -ctl /home/mcc/execution/CTLFireability.ctl
No direction supplied, using forward translation only.
Parsed 16 CTL formulae.
built 17 ordering constraints for composite.
built 15 ordering constraints for composite.
built 8 ordering constraints for composite.
built 20 ordering constraints for composite.
built 14 ordering constraints for composite.
built 24 ordering constraints for composite.
Model ,|S| ,Time ,Mem(kb) ,fin. SDD ,fin. DDD ,peak SDD ,peak DDD ,SDD Hom ,SDD cache peak ,DDD Hom ,DDD cachepeak ,SHom cache
reachable,4.27349e+07,8.3801,104828,2673,490,164982,7114,217,284083,59,67542,0
Converting to forward existential form...Done !
original formula: !(((AF((((i0.u3.KdStarGStar>=1)&&(i2.u4.P3k>=1))&&(i0.u3.KdStarG>=1))) + AG(!((i4.u10.KdStarGStarPgP3>=1)))) + (i4.u8.KdStarGStarPgStarP3P2>=1)))
=> equivalent forward existential formula: [(FwdU(((Init * !((i4.u8.KdStarGStarPgStarP3P2>=1))) * !(!(EG(!((((i0.u3.KdStarGStar>=1)&&(i2.u4.P3k>=1))&&(i0.u3.KdStarG>=1))))))),TRUE) * (i4.u10.KdStarGStarPgP3>=1))] != FALSE
Reverse transition relation is NOT exact ! Due to transitions k55, i0.u3.k2, i2.u4.k7, i5.u12.k60, Intersection with reachable at each step enabled. (destroyed/reverse/intersect/total) :0/60/4/64
Fast SCC detection found an SCC at level 1
Fast SCC detection found a local SCC at level 1
Fast SCC detection found an SCC at level 2
Fast SCC detection found an SCC at level 3
Fast SCC detection found an SCC at level 4
Fast SCC detection found an SCC at level 5
(forward)formula 0,1,44.6187,492712,1,0,712673,58855,1104,1.70262e+06,388,436375,936505
FORMULA Angiogenesis-PT-05-CTLFireability-00 TRUE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL USE_NUPN
Formula is TRUE !
***************************************
original formula: (EF(AG((((i0.u7.KdStarGStarPgStar>=1)&&(i5.u12.Pip2>=1))&&((i2.u4.GStarP3>=1)&&(i2.u4.KdStar>=1))))) + AG(AX((i5.u12.PtP2>=1))))
=> equivalent forward existential formula: [(EY(FwdU((Init * !(E(TRUE U !(E(TRUE U !((((i0.u7.KdStarGStarPgStar>=1)&&(i5.u12.Pip2>=1))&&((i2.u4.GStarP3>=1)&&(i2.u4.KdStar>=1))))))))),TRUE)) * !((i5.u12.PtP2>=1)))] = FALSE
(forward)formula 1,0,45.3135,498784,1,0,718957,59156,1234,1.7241e+06,399,440152,963564
FORMULA Angiogenesis-PT-05-CTLFireability-01 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL USE_NUPN
Formula is FALSE !
***************************************
original formula: (EF((AX((i4.u8.KdStarGStarPgStarP3P2>=1)) * (((i2.u4.GStarP3>=1)&&(i4.u10.Pg>=1))&&((i5.u12.PtP3P2>=1)||(i2.u6.KdStarGStarP3kStarP3P2>=1))))) * AF(EF(((i2.u6.KdStarGStarP3kStarP3P2>=1)&&(i1.u9.KdStarPg>=1)))))
=> equivalent forward existential formula: ([(Init * !(E(TRUE U (!(EX(!((i4.u8.KdStarGStarPgStarP3P2>=1)))) * (((i2.u4.GStarP3>=1)&&(i4.u10.Pg>=1))&&((i5.u12.PtP3P2>=1)||(i2.u6.KdStarGStarP3kStarP3P2>=1)))))))] = FALSE * [FwdG(Init,!(E(TRUE U ((i2.u6.KdStarGStarP3kStarP3P2>=1)&&(i1.u9.KdStarPg>=1)))))] = FALSE)
(forward)formula 2,0,54.6019,581952,1,0,851177,59999,1249,1.8945e+06,399,448232,1197872
FORMULA Angiogenesis-PT-05-CTLFireability-02 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL USE_NUPN
Formula is FALSE !
***************************************
original formula: AG(EF(EG((i1.u1.DAGE>=1))))
=> equivalent forward existential formula: [(FwdU(Init,TRUE) * !(E(TRUE U EG((i1.u1.DAGE>=1)))))] = FALSE
(forward)formula 3,0,56.6293,607032,1,0,899638,60089,1262,1.93511e+06,399,448565,1259330
FORMULA Angiogenesis-PT-05-CTLFireability-03 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL USE_NUPN
Formula is FALSE !
***************************************
original formula: !(EF(AG((i5.u0.AktP3>=1))))
=> equivalent forward existential formula: [(FwdU(Init,TRUE) * !(E(TRUE U !((i5.u0.AktP3>=1)))))] = FALSE
(forward)formula 4,1,56.6432,607560,1,0,899746,60089,1265,1.93524e+06,399,448567,1259521
FORMULA Angiogenesis-PT-05-CTLFireability-04 TRUE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL USE_NUPN
Formula is TRUE !
***************************************
original formula: ((i5.u11.PtP3>=1) + (((!(i5.u11.PtP3>=1))&&((i5.u0.Pip3>=1)&&(i5.u11.Pten>=1))) + ((((i2.u4.GStarP3kP3>=1)&&(i2.u4.KdStar>=1)) * AX((i0.u7.KdStarGStarPg>=1))) + (EF(((i4.u8.KdStarGStarPgStarP3>=1)&&(i5.u12.Pip2>=1))) + ((i2.u4.GStarP3>=1)&&(i2.u4.KdStar>=1))))))
=> equivalent forward existential formula: ([((((Init * !((i5.u11.PtP3>=1))) * !(((!(i5.u11.PtP3>=1))&&((i5.u0.Pip3>=1)&&(i5.u11.Pten>=1))))) * !((E(TRUE U ((i4.u8.KdStarGStarPgStarP3>=1)&&(i5.u12.Pip2>=1))) + ((i2.u4.GStarP3>=1)&&(i2.u4.KdStar>=1))))) * !(((i2.u4.GStarP3kP3>=1)&&(i2.u4.KdStar>=1))))] = FALSE * [(EY((((Init * !((i5.u11.PtP3>=1))) * !(((!(i5.u11.PtP3>=1))&&((i5.u0.Pip3>=1)&&(i5.u11.Pten>=1))))) * !((E(TRUE U ((i4.u8.KdStarGStarPgStarP3>=1)&&(i5.u12.Pip2>=1))) + ((i2.u4.GStarP3>=1)&&(i2.u4.KdStar>=1)))))) * !((i0.u7.KdStarGStarPg>=1)))] = FALSE)
(forward)formula 5,1,60.1264,640560,1,0,941527,60492,1299,2.02034e+06,405,451900,1327307
FORMULA Angiogenesis-PT-05-CTLFireability-05 TRUE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL USE_NUPN
Formula is TRUE !
***************************************
original formula: E(!(((i2.u6.KdStarGStarP3kStarP3>=1)&&(i5.u12.Pip2>=1))) U ((i0.u7.KdStarGStarPgStarP2>=1)&&((i2.u4.GStarP3kP3>=1)&&(i2.u4.KdStar>=1))))
=> equivalent forward existential formula: [(FwdU(Init,!(((i2.u6.KdStarGStarP3kStarP3>=1)&&(i5.u12.Pip2>=1)))) * ((i0.u7.KdStarGStarPgStarP2>=1)&&((i2.u4.GStarP3kP3>=1)&&(i2.u4.KdStar>=1))))] != FALSE
(forward)formula 6,1,182.606,1742888,1,0,2.11899e+06,98918,1326,8.369e+06,405,1.41018e+06,2516293
FORMULA Angiogenesis-PT-05-CTLFireability-06 TRUE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL USE_NUPN
Formula is TRUE !
***************************************
original formula: (E(((!((i2.u4.GStarP3>=1)&&(i2.u4.P3k>=1)))||(((i1.u9.KdStarPgStar>=1)&&(i5.u12.Pip2>=1))||((i2.u4.GStarP3kP3>=1)&&(i2.u4.KdStar>=1)))) U (((i2.u4.KdStarGP3>=1)&&(i0.u2.GP3>=1))&&(!(i2.u4.KdStarGP3>=1)))) + (((i2.u4.GStarP3>=1)&&(i2.u4.P3k>=1)) + ((EX((i4.u8.KdStarGStarPgStarP3P2>=1)) + AX((i4.u10.KdStarGStarPgP3>=1))) + AF(((i5.u0.AktP3>=1)&&(i5.u11.PtP3>=1))))))
=> equivalent forward existential formula: [FwdG((((Init * !(E(((!((i2.u4.GStarP3>=1)&&(i2.u4.P3k>=1)))||(((i1.u9.KdStarPgStar>=1)&&(i5.u12.Pip2>=1))||((i2.u4.GStarP3kP3>=1)&&(i2.u4.KdStar>=1)))) U (((i2.u4.KdStarGP3>=1)&&(i0.u2.GP3>=1))&&(!(i2.u4.KdStarGP3>=1)))))) * !(((i2.u4.GStarP3>=1)&&(i2.u4.P3k>=1)))) * !((EX((i4.u8.KdStarGStarPgStarP3P2>=1)) + !(EX(!((i4.u10.KdStarGStarPgP3>=1))))))),!(((i5.u0.AktP3>=1)&&(i5.u11.PtP3>=1))))] = FALSE
(forward)formula 7,0,200.313,1920032,1,0,2.32007e+06,104189,1392,9.08935e+06,412,1.57142e+06,2782291
FORMULA Angiogenesis-PT-05-CTLFireability-07 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL USE_NUPN
Formula is FALSE !
***************************************
original formula: A((i0.u7.KdStarGStarPg>=1) U EF(((i4.u10.KdStarGStarPgP3>=1)&&(i2.u4.GStarP3kP3>=1))))
=> equivalent forward existential formula: [((Init * !(EG(!(E(TRUE U ((i4.u10.KdStarGStarPgP3>=1)&&(i2.u4.GStarP3kP3>=1))))))) * !(E(!(E(TRUE U ((i4.u10.KdStarGStarPgP3>=1)&&(i2.u4.GStarP3kP3>=1)))) U (!((i0.u7.KdStarGStarPg>=1)) * !(E(TRUE U ((i4.u10.KdStarGStarPgP3>=1)&&(i2.u4.GStarP3kP3>=1))))))))] != FALSE
Fast SCC detection found an SCC at level 1
Fast SCC detection found a local SCC at level 2
Fast SCC detection found an SCC at level 3
Fast SCC detection found an SCC at level 4
Fast SCC detection found an SCC at level 5
(forward)formula 8,1,213.453,2013752,1,0,2.4449e+06,104549,1425,9.27838e+06,416,1.57398e+06,3009960
FORMULA Angiogenesis-PT-05-CTLFireability-08 TRUE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL USE_NUPN
Formula is TRUE !
***************************************
original formula: (!(EF(EX((i2.u4.KdStarGStarP3kP3>=1)))) + EF((((!((i2.u4.GStarP3>=1)&&(i2.u4.KdStar>=1)))||((i5.u12.PtP2>=1)||(i0.u3.KdStarG>=1)))&&((((i0.u2.GP3>=1)&&(i2.u4.KdStar>=1))||((i0.u2.Gab1>=1)&&(i2.u4.KdStar>=1)))&&((i4.u8.KdStarGStarPgStarP3P2>=1)||(i2.u4.GStarP3kP3>=1))))))
=> equivalent forward existential formula: [(EY(FwdU((Init * !(E(TRUE U (((!((i2.u4.GStarP3>=1)&&(i2.u4.KdStar>=1)))||((i5.u12.PtP2>=1)||(i0.u3.KdStarG>=1)))&&((((i0.u2.GP3>=1)&&(i2.u4.KdStar>=1))||((i0.u2.Gab1>=1)&&(i2.u4.KdStar>=1)))&&((i4.u8.KdStarGStarPgStarP3P2>=1)||(i2.u4.GStarP3kP3>=1))))))),TRUE)) * (i2.u4.KdStarGStarP3kP3>=1))] = FALSE
(forward)formula 9,1,220.805,2062064,1,0,2.51199e+06,105403,1450,9.39505e+06,418,1.58663e+06,3126131
FORMULA Angiogenesis-PT-05-CTLFireability-09 TRUE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL USE_NUPN
Formula is TRUE !
***************************************
original formula: AG(EF(((i0.u2.Gab1>=1)&&(i2.u4.KdStar>=1))))
=> equivalent forward existential formula: [(FwdU(Init,TRUE) * !(E(TRUE U ((i0.u2.Gab1>=1)&&(i2.u4.KdStar>=1)))))] = FALSE
(forward)formula 10,0,223.761,2081864,1,0,2.54751e+06,105408,1450,9.42497e+06,418,1.58668e+06,3183012
FORMULA Angiogenesis-PT-05-CTLFireability-10 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL USE_NUPN
Formula is FALSE !
***************************************
original formula: !(EF((i4.u10.KdStarGStarPgP3>=1)))
=> equivalent forward existential formula: [(FwdU(Init,TRUE) * (i4.u10.KdStarGStarPgP3>=1))] = FALSE
(forward)formula 11,0,223.763,2082128,1,0,2.54751e+06,105408,1450,9.42497e+06,418,1.58668e+06,3183012
FORMULA Angiogenesis-PT-05-CTLFireability-11 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL USE_NUPN
Formula is FALSE !
***************************************
original formula: ((((!(((i0.u2.Gab1>=1)&&(i5.u0.Pip3>=1))&&(!((u5.KdStarGStarP3kStar>=1)&&(i5.u12.Pip2>=1)))))||((i5.u12.PtP2>=1)&&(i5.u0.Pip3>=1))) * (E((i2.u6.KdStarGStarP3kStarP3P2>=1) U (i4.u8.KdStarGStarPgStarP3P2>=1)) + !(((i1.u9.KdStarPgStarP2>=1)&&(!((i2.u4.KdStar>=1)&&(i4.u10.Pg>=1))))))) * AG(!((i2.u6.KdStarGStarP3kStarP3P2>=1))))
=> equivalent forward existential formula: (([(Init * !(((!(((i0.u2.Gab1>=1)&&(i5.u0.Pip3>=1))&&(!((u5.KdStarGStarP3kStar>=1)&&(i5.u12.Pip2>=1)))))||((i5.u12.PtP2>=1)&&(i5.u0.Pip3>=1)))))] = FALSE * [((Init * !(!(((i1.u9.KdStarPgStarP2>=1)&&(!((i2.u4.KdStar>=1)&&(i4.u10.Pg>=1))))))) * !(E((i2.u6.KdStarGStarP3kStarP3P2>=1) U (i4.u8.KdStarGStarPgStarP3P2>=1))))] = FALSE) * [(FwdU(Init,TRUE) * (i2.u6.KdStarGStarP3kStarP3P2>=1))] = FALSE)
(forward)formula 12,0,223.773,2082128,1,0,2.54751e+06,105408,1450,9.42522e+06,418,1.58668e+06,3183013
FORMULA Angiogenesis-PT-05-CTLFireability-12 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL USE_NUPN
Formula is FALSE !
***************************************
original formula: AG(AX(EF((i5.u12.PtP2>=1))))
=> equivalent forward existential formula: [(EY(FwdU(Init,TRUE)) * !(E(TRUE U (i5.u12.PtP2>=1))))] = FALSE
(forward)formula 13,0,223.871,2082392,1,0,2.54839e+06,105408,1450,9.42617e+06,418,1.58668e+06,3185396
FORMULA Angiogenesis-PT-05-CTLFireability-13 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL USE_NUPN
Formula is FALSE !
***************************************
original formula: E(EF((i2.u4.KdStarGP3>=1)) U (((i2.u4.GStarP3>=1)&&(i2.u4.P3k>=1)) * EG(((i2.u4.GStarP3>=1)&&(i4.u10.Pg>=1)))))
=> equivalent forward existential formula: [FwdG((FwdU(Init,E(TRUE U (i2.u4.KdStarGP3>=1))) * ((i2.u4.GStarP3>=1)&&(i2.u4.P3k>=1))),((i2.u4.GStarP3>=1)&&(i4.u10.Pg>=1)))] != FALSE
(forward)formula 14,1,343.204,2205284,1,0,2.75555e+06,111815,617,9.88466e+06,192,1.6269e+06,664759
FORMULA Angiogenesis-PT-05-CTLFireability-14 TRUE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL USE_NUPN
Formula is TRUE !
***************************************
original formula: ((!(EF((i2.u6.KdStarGStarP3kStarP3P2>=1))) + ((((i1.u9.KdStarPgStar>=1)&&(i5.u12.Pip2>=1))&&((i4.u10.GStarPgP3>=1)&&((i5.u11.PtP3>=1)||(i2.u4.KdStarGStarP3kP3>=1)))) + AX((((i2.u4.GStarP3kP3>=1)&&(i2.u4.KdStar>=1))||(i0.u3.KdStarG>=1))))) * !(EG(EX((i1.u1.DAGE>=1)))))
=> equivalent forward existential formula: ([(EY(((Init * !(!(E(TRUE U (i2.u6.KdStarGStarP3kStarP3P2>=1))))) * !((((i1.u9.KdStarPgStar>=1)&&(i5.u12.Pip2>=1))&&((i4.u10.GStarPgP3>=1)&&((i5.u11.PtP3>=1)||(i2.u4.KdStarGStarP3kP3>=1))))))) * !((((i2.u4.GStarP3kP3>=1)&&(i2.u4.KdStar>=1))||(i0.u3.KdStarG>=1))))] = FALSE * [FwdG(Init,EX((i1.u1.DAGE>=1)))] = FALSE)
(forward)formula 15,0,349.71,2205284,1,0,2.75555e+06,111815,914,9.88466e+06,226,1.6269e+06,953403
FORMULA Angiogenesis-PT-05-CTLFireability-15 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL USE_NUPN
Formula is FALSE !
***************************************
BK_STOP 1527201068694
--------------------
content from stderr:
+ export BINDIR=/home/mcc/BenchKit/
+ BINDIR=/home/mcc/BenchKit/
++ pwd
+ export MODEL=/home/mcc/execution
+ MODEL=/home/mcc/execution
+ /home/mcc/BenchKit//runeclipse.sh /home/mcc/execution CTLFireability -its -ltsminpath /home/mcc/BenchKit//lts_install_dir/ -louvain -smt
+ ulimit -s 65536
+ [[ -z '' ]]
+ export LTSMIN_MEM_SIZE=8589934592
+ LTSMIN_MEM_SIZE=8589934592
+ /home/mcc/BenchKit//itstools/its-tools -consoleLog -data /home/mcc/execution/workspace -pnfolder /home/mcc/execution -examination CTLFireability -z3path /home/mcc/BenchKit//z3/bin/z3 -yices2path /home/mcc/BenchKit//yices/bin/yices -its -ltsminpath /home/mcc/BenchKit//lts_install_dir/ -louvain -smt -vmargs -Dosgi.locking=none -Declipse.stateSaveDelayInterval=-1 -Dosgi.configuration.area=/tmp/.eclipse -Xss8m -Xms40m -Xmx8192m -Dfile.encoding=UTF-8 -Dosgi.requiredJavaVersion=1.6
May 24, 2018 10:25:15 PM fr.lip6.move.gal.application.Application start
INFO: Running its-tools with arguments : [-pnfolder, /home/mcc/execution, -examination, CTLFireability, -z3path, /home/mcc/BenchKit//z3/bin/z3, -yices2path, /home/mcc/BenchKit//yices/bin/yices, -its, -ltsminpath, /home/mcc/BenchKit//lts_install_dir/, -louvain, -smt]
May 24, 2018 10:25:15 PM fr.lip6.move.gal.application.MccTranslator transformPNML
INFO: Parsing pnml file : /home/mcc/execution/model.pnml
May 24, 2018 10:25:15 PM fr.lip6.move.gal.nupn.PTNetReader loadFromXML
INFO: Load time of PNML (sax parser for PT used): 58 ms
May 24, 2018 10:25:15 PM fr.lip6.move.gal.pnml.togal.PTGALTransformer handlePage
INFO: Transformed 39 places.
May 24, 2018 10:25:16 PM fr.lip6.move.gal.pnml.togal.PTGALTransformer handlePage
INFO: Transformed 64 transitions.
May 24, 2018 10:25:16 PM fr.lip6.move.serialization.SerializationUtil systemToFile
INFO: Time to serialize gal into /home/mcc/execution/model.pnml.img.gal : 17 ms
May 24, 2018 10:25:16 PM fr.lip6.move.gal.application.MccTranslator applyOrder
INFO: Applying decomposition
May 24, 2018 10:25:16 PM fr.lip6.move.gal.instantiate.GALRewriter flatten
INFO: Flatten gal took : 61 ms
May 24, 2018 10:25:16 PM fr.lip6.move.gal.instantiate.GALRewriter flatten
INFO: Flatten gal took : 23 ms
Begin: Thu May 24 22:25:16 2018
Computation of communities with the Newman-Girvan Modularity quality function
level 0:
start computation: Thu May 24 22:25:16 2018
network size: 39 nodes, 138 links, 128 weight
quality increased from -0.0310669 to 0.427765
end computation: Thu May 24 22:25:16 2018
level 1:
start computation: Thu May 24 22:25:16 2018
network size: 13 nodes, 69 links, 128 weight
quality increased from 0.427765 to 0.482574
end computation: Thu May 24 22:25:16 2018
level 2:
start computation: Thu May 24 22:25:16 2018
network size: 6 nodes, 32 links, 128 weight
quality increased from 0.482574 to 0.482574
end computation: Thu May 24 22:25:16 2018
End: Thu May 24 22:25:16 2018
Total duration: 0 sec
0.482574
May 24, 2018 10:25:16 PM fr.lip6.move.gal.instantiate.CompositeBuilder decomposeWithOrder
INFO: Decomposing Gal with order
May 24, 2018 10:25:16 PM fr.lip6.move.gal.instantiate.GALRewriter flatten
INFO: Flatten gal took : 19 ms
May 24, 2018 10:25:16 PM fr.lip6.move.gal.instantiate.CompositeBuilder rewriteArraysToAllowPartition
INFO: Rewriting arrays to variables to allow decomposition.
May 24, 2018 10:25:16 PM fr.lip6.move.gal.instantiate.Instantiator fuseIsomorphicEffects
INFO: Removed a total of 33 redundant transitions.
May 24, 2018 10:25:16 PM fr.lip6.move.serialization.SerializationUtil systemToFile
INFO: Time to serialize gal into /home/mcc/execution/CTLFireability.pnml.gal : 5 ms
May 24, 2018 10:25:16 PM fr.lip6.move.serialization.SerializationUtil serializePropertiesForITSCTLTools
INFO: Time to serialize properties into /home/mcc/execution/CTLFireability.ctl : 2 ms
Sequence of Actions to be Executed by the VM
This is useful if one wants to reexecute the tool in the VM from the submitted image disk.
set -x
# this is for BenchKit: configuration of major elements for the test
export BK_INPUT="Angiogenesis-PT-05"
export BK_EXAMINATION="CTLFireability"
export BK_TOOL="itstoolsl"
export BK_RESULT_DIR="/tmp/BK_RESULTS/OUTPUTS"
export BK_TIME_CONFINEMENT="3600"
export BK_MEMORY_CONFINEMENT="16384"
# this is specific to your benchmark or test
export BIN_DIR="$HOME/BenchKit/bin"
# remove the execution directoty if it exists (to avoid increse of .vmdk images)
if [ -d execution ] ; then
rm -rf execution
fi
tar xzf /home/mcc/BenchKit/INPUTS/Angiogenesis-PT-05.tgz
mv Angiogenesis-PT-05 execution
cd execution
pwd
ls -lh
# this is for BenchKit: explicit launching of the test
echo "====================================================================="
echo " Generated by BenchKit 2-3637"
echo " Executing tool itstoolsl"
echo " Input is Angiogenesis-PT-05, examination is CTLFireability"
echo " Time confinement is $BK_TIME_CONFINEMENT seconds"
echo " Memory confinement is 16384 MBytes"
echo " Number of cores is 4"
echo " Run identifier is r021-qhx1-152646246200011"
echo "====================================================================="
echo
echo "--------------------"
echo "content from stdout:"
echo
echo "=== Data for post analysis generated by BenchKit (invocation template)"
echo
if [ "CTLFireability" = "UpperBounds" ] ; then
echo "The expected result is a vector of positive values"
echo NUM_VECTOR
elif [ "CTLFireability" != "StateSpace" ] ; then
echo "The expected result is a vector of booleans"
echo BOOL_VECTOR
else
echo "no data necessary for post analysis"
fi
echo
if [ -f "CTLFireability.txt" ] ; then
echo "here is the order used to build the result vector(from text file)"
for x in $(grep Property CTLFireability.txt | cut -d ' ' -f 2 | sort -u) ; do
echo "FORMULA_NAME $x"
done
elif [ -f "CTLFireability.xml" ] ; then # for cunf (txt files deleted;-)
echo echo "here is the order used to build the result vector(from xml file)"
for x in $(grep '
echo "FORMULA_NAME $x"
done
fi
echo
echo "=== Now, execution of the tool begins"
echo
echo -n "BK_START "
date -u +%s%3N
echo
timeout -s 9 $BK_TIME_CONFINEMENT bash -c "/home/mcc/BenchKit/BenchKit_head.sh 2> STDERR ; echo ; echo -n \"BK_STOP \" ; date -u +%s%3N"
if [ $? -eq 137 ] ; then
echo
echo "BK_TIME_CONFINEMENT_REACHED"
fi
echo
echo "--------------------"
echo "content from stderr:"
echo
cat STDERR ;

