About the Execution of LTSMin for Angiogenesis-PT-05
| Execution Summary | |||||
| Max Memory Used (MB) |
Time wait (ms) | CPU Usage (ms) | I/O Wait (ms) | Computed Result | Execution Status |
| 9385.470 | 90312.00 | 322419.00 | 1454.80 | 2 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 | normal |
Execution Chart
We display below the execution chart for this examination (boot time has been removed).
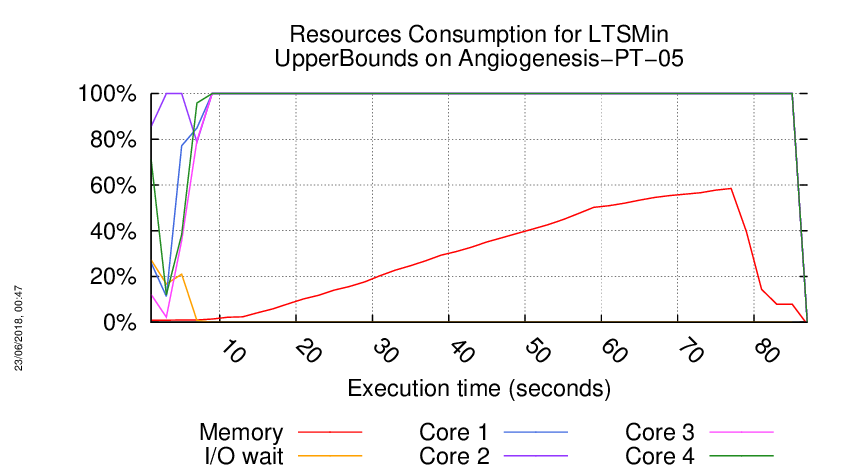
Trace from the execution
Waiting for the VM to be ready (probing ssh)
.........................
/home/mcc/execution
total 204K
-rw-r--r-- 1 mcc users 3.1K May 15 18:54 CTLCardinality.txt
-rw-r--r-- 1 mcc users 15K May 15 18:54 CTLCardinality.xml
-rw-r--r-- 1 mcc users 2.6K May 15 18:54 CTLFireability.txt
-rw-r--r-- 1 mcc users 19K May 15 18:54 CTLFireability.xml
-rw-r--r-- 1 mcc users 4.0K May 15 18:49 GenericPropertiesDefinition.xml
-rw-r--r-- 1 mcc users 6.0K May 15 18:49 GenericPropertiesVerdict.xml
-rw-r--r-- 1 mcc users 2.5K May 15 18:54 LTLCardinality.txt
-rw-r--r-- 1 mcc users 11K May 15 18:54 LTLCardinality.xml
-rw-r--r-- 1 mcc users 2.0K May 15 18:54 LTLFireability.txt
-rw-r--r-- 1 mcc users 11K May 15 18:54 LTLFireability.xml
-rw-r--r-- 1 mcc users 4.4K May 15 18:54 ReachabilityCardinality.txt
-rw-r--r-- 1 mcc users 21K May 15 18:54 ReachabilityCardinality.xml
-rw-r--r-- 1 mcc users 109 May 15 18:54 ReachabilityDeadlock.txt
-rw-r--r-- 1 mcc users 347 May 15 18:54 ReachabilityDeadlock.xml
-rw-r--r-- 1 mcc users 2.4K May 15 18:54 ReachabilityFireability.txt
-rw-r--r-- 1 mcc users 14K May 15 18:54 ReachabilityFireability.xml
-rw-r--r-- 1 mcc users 1.7K May 15 18:54 UpperBounds.txt
-rw-r--r-- 1 mcc users 3.8K May 15 18:54 UpperBounds.xml
-rw-r--r-- 1 mcc users 6 May 15 18:49 equiv_col
-rw-r--r-- 1 mcc users 3 May 15 18:49 instance
-rw-r--r-- 1 mcc users 6 May 15 18:49 iscolored
-rw-r--r-- 1 mcc users 33K May 15 18:49 model.pnml
=====================================================================
Generated by BenchKit 2-3637
Executing tool ltsmin
Input is Angiogenesis-PT-05, examination is UpperBounds
Time confinement is 3600 seconds
Memory confinement is 16384 MBytes
Number of cores is 4
Run identifier is r017-qhx1-152646241800009
=====================================================================
--------------------
content from stdout:
=== Data for post analysis generated by BenchKit (invocation template)
The expected result is a vector of positive values
NUM_VECTOR
here is the order used to build the result vector(from text file)
FORMULA_NAME Angiogenesis-PT-05-UpperBounds-00
FORMULA_NAME Angiogenesis-PT-05-UpperBounds-01
FORMULA_NAME Angiogenesis-PT-05-UpperBounds-02
FORMULA_NAME Angiogenesis-PT-05-UpperBounds-03
FORMULA_NAME Angiogenesis-PT-05-UpperBounds-04
FORMULA_NAME Angiogenesis-PT-05-UpperBounds-05
FORMULA_NAME Angiogenesis-PT-05-UpperBounds-06
FORMULA_NAME Angiogenesis-PT-05-UpperBounds-07
FORMULA_NAME Angiogenesis-PT-05-UpperBounds-08
FORMULA_NAME Angiogenesis-PT-05-UpperBounds-09
FORMULA_NAME Angiogenesis-PT-05-UpperBounds-10
FORMULA_NAME Angiogenesis-PT-05-UpperBounds-11
FORMULA_NAME Angiogenesis-PT-05-UpperBounds-12
FORMULA_NAME Angiogenesis-PT-05-UpperBounds-13
FORMULA_NAME Angiogenesis-PT-05-UpperBounds-14
FORMULA_NAME Angiogenesis-PT-05-UpperBounds-15
=== Now, execution of the tool begins
BK_START 1526683426018
FORMULA Angiogenesis-PT-05-UpperBounds-00 2 TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN
FORMULA Angiogenesis-PT-05-UpperBounds-01 5 TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN
FORMULA Angiogenesis-PT-05-UpperBounds-02 5 TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN
FORMULA Angiogenesis-PT-05-UpperBounds-03 5 TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN
FORMULA Angiogenesis-PT-05-UpperBounds-04 5 TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN
FORMULA Angiogenesis-PT-05-UpperBounds-05 5 TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN
FORMULA Angiogenesis-PT-05-UpperBounds-06 5 TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN
FORMULA Angiogenesis-PT-05-UpperBounds-07 5 TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN
FORMULA Angiogenesis-PT-05-UpperBounds-08 5 TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN
FORMULA Angiogenesis-PT-05-UpperBounds-09 5 TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN
FORMULA Angiogenesis-PT-05-UpperBounds-10 5 TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN
FORMULA Angiogenesis-PT-05-UpperBounds-11 5 TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN
FORMULA Angiogenesis-PT-05-UpperBounds-12 5 TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN
FORMULA Angiogenesis-PT-05-UpperBounds-13 5 TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN
FORMULA Angiogenesis-PT-05-UpperBounds-14 5 TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN
FORMULA Angiogenesis-PT-05-UpperBounds-15 5 TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN
BK_STOP 1526683516330
--------------------
content from stderr:
mcc2018
ub formula name Angiogenesis-PT-05-UpperBounds-00
ub formula formula --maxsum=/tmp/ub_0_
ub formula name Angiogenesis-PT-05-UpperBounds-01
ub formula formula --maxsum=/tmp/ub_1_
ub formula name Angiogenesis-PT-05-UpperBounds-02
ub formula formula --maxsum=/tmp/ub_2_
ub formula name Angiogenesis-PT-05-UpperBounds-03
ub formula formula --maxsum=/tmp/ub_3_
ub formula name Angiogenesis-PT-05-UpperBounds-04
ub formula formula --maxsum=/tmp/ub_4_
ub formula name Angiogenesis-PT-05-UpperBounds-05
ub formula formula --maxsum=/tmp/ub_5_
ub formula name Angiogenesis-PT-05-UpperBounds-06
ub formula formula --maxsum=/tmp/ub_6_
ub formula name Angiogenesis-PT-05-UpperBounds-07
ub formula formula --maxsum=/tmp/ub_7_
ub formula name Angiogenesis-PT-05-UpperBounds-08
ub formula formula --maxsum=/tmp/ub_8_
ub formula name Angiogenesis-PT-05-UpperBounds-09
ub formula formula --maxsum=/tmp/ub_9_
ub formula name Angiogenesis-PT-05-UpperBounds-10
ub formula formula --maxsum=/tmp/ub_10_
ub formula name Angiogenesis-PT-05-UpperBounds-11
ub formula formula --maxsum=/tmp/ub_11_
ub formula name Angiogenesis-PT-05-UpperBounds-12
ub formula formula --maxsum=/tmp/ub_12_
ub formula name Angiogenesis-PT-05-UpperBounds-13
ub formula formula --maxsum=/tmp/ub_13_
ub formula name Angiogenesis-PT-05-UpperBounds-14
ub formula formula --maxsum=/tmp/ub_14_
ub formula name Angiogenesis-PT-05-UpperBounds-15
ub formula formula --maxsum=/tmp/ub_15_
pnml2lts-sym: Exploration order is chain-prev
pnml2lts-sym: Saturation strategy is sat-like
pnml2lts-sym: Guided search strategy is unguided
pnml2lts-sym: Attractor strategy is default
pnml2lts-sym: opening model.pnml
pnml2lts-sym: Edge label is id
pnml2lts-sym: Petri net has 39 places, 64 transitions and 185 arcs
pnml2lts-sym: Petri net Angiogenesis-PT-05 analyzed
pnml2lts-sym: There are no safe places
pnml2lts-sym: Loading Petri net took 0.000 real 0.000 user 0.000 sys
pnml2lts-sym: Initializing regrouping layer
pnml2lts-sym: Regroup specification: bs,w2W,ru,hf
pnml2lts-sym: Regroup Boost's Sloan
pnml2lts-sym: Regroup over-approximate must-write to may-write
pnml2lts-sym: Regroup Row sUbsume
pnml2lts-sym: Reqroup Horizontal Flip
pnml2lts-sym: Regrouping: 64->40 groups
pnml2lts-sym: Regrouping took 0.000 real 0.000 user 0.000 sys
pnml2lts-sym: state vector length is 39; there are 40 groups
pnml2lts-sym: Computing maximum sum over 1 state variables.
pnml2lts-sym: Computing maximum sum over 1 state variables.
pnml2lts-sym: Computing maximum sum over 1 state variables.
pnml2lts-sym: Computing maximum sum over 1 state variables.
pnml2lts-sym: Computing maximum sum over 1 state variables.
pnml2lts-sym: Computing maximum sum over 1 state variables.
pnml2lts-sym: Computing maximum sum over 1 state variables.
pnml2lts-sym: Computing maximum sum over 1 state variables.
pnml2lts-sym: Computing maximum sum over 1 state variables.
pnml2lts-sym: Computing maximum sum over 1 state variables.
pnml2lts-sym: Computing maximum sum over 1 state variables.
pnml2lts-sym: Computing maximum sum over 1 state variables.
pnml2lts-sym: Computing maximum sum over 1 state variables.
pnml2lts-sym: Computing maximum sum over 1 state variables.
pnml2lts-sym: Computing maximum sum over 1 state variables.
pnml2lts-sym: Computing maximum sum over 1 state variables.
pnml2lts-sym: Creating a multi-core ListDD domain.
pnml2lts-sym: Sylvan allocates 15.000 GB virtual memory for nodes table and operation cache.
pnml2lts-sym: Initial nodes table and operation cache requires 15.000 GB.
pnml2lts-sym: Using GBgetTransitionsShortR2W as next-state function
pnml2lts-sym: got initial state
pnml2lts-sym: Exploration took 1678 group checks and 1678 next state calls
pnml2lts-sym: reachability took 76.330 real 138.460 user 162.580 sys
pnml2lts-sym: counting visited states...
pnml2lts-sym: counting took 0.270 real 0.540 user 0.560 sys
pnml2lts-sym: state space has 42734935 states, 10829 nodes
pnml2lts-sym: Maximum sum of /tmp/ub_0_ is: 2
pnml2lts-sym: Maximum sum of /tmp/ub_1_ is: 5
pnml2lts-sym: Maximum sum of /tmp/ub_2_ is: 5
pnml2lts-sym: Maximum sum of /tmp/ub_3_ is: 5
pnml2lts-sym: Maximum sum of /tmp/ub_4_ is: 5
pnml2lts-sym: Maximum sum of /tmp/ub_5_ is: 5
pnml2lts-sym: Maximum sum of /tmp/ub_6_ is: 5
pnml2lts-sym: Maximum sum of /tmp/ub_7_ is: 5
pnml2lts-sym: Maximum sum of /tmp/ub_8_ is: 5
pnml2lts-sym: Maximum sum of /tmp/ub_9_ is: 5
pnml2lts-sym: Maximum sum of /tmp/ub_10_ is: 5
pnml2lts-sym: Maximum sum of /tmp/ub_11_ is: 5
pnml2lts-sym: Maximum sum of /tmp/ub_12_ is: 5
pnml2lts-sym: Maximum sum of /tmp/ub_13_ is: 5
pnml2lts-sym: Maximum sum of /tmp/ub_14_ is: 5
pnml2lts-sym: Maximum sum of /tmp/ub_15_ is: 5
pnml2lts-sym: group_next: 3014 nodes total
pnml2lts-sym: group_explored: 1656 nodes, 2350 short vectors total
pnml2lts-sym: max token count: 5
Sequence of Actions to be Executed by the VM
This is useful if one wants to reexecute the tool in the VM from the submitted image disk.
set -x
# this is for BenchKit: configuration of major elements for the test
export BK_INPUT="Angiogenesis-PT-05"
export BK_EXAMINATION="UpperBounds"
export BK_TOOL="ltsmin"
export BK_RESULT_DIR="/tmp/BK_RESULTS/OUTPUTS"
export BK_TIME_CONFINEMENT="3600"
export BK_MEMORY_CONFINEMENT="16384"
# this is specific to your benchmark or test
export BIN_DIR="$HOME/BenchKit/bin"
# remove the execution directoty if it exists (to avoid increse of .vmdk images)
if [ -d execution ] ; then
rm -rf execution
fi
tar xzf /home/mcc/BenchKit/INPUTS/Angiogenesis-PT-05.tgz
mv Angiogenesis-PT-05 execution
cd execution
pwd
ls -lh
# this is for BenchKit: explicit launching of the test
echo "====================================================================="
echo " Generated by BenchKit 2-3637"
echo " Executing tool ltsmin"
echo " Input is Angiogenesis-PT-05, examination is UpperBounds"
echo " Time confinement is $BK_TIME_CONFINEMENT seconds"
echo " Memory confinement is 16384 MBytes"
echo " Number of cores is 4"
echo " Run identifier is r017-qhx1-152646241800009"
echo "====================================================================="
echo
echo "--------------------"
echo "content from stdout:"
echo
echo "=== Data for post analysis generated by BenchKit (invocation template)"
echo
if [ "UpperBounds" = "UpperBounds" ] ; then
echo "The expected result is a vector of positive values"
echo NUM_VECTOR
elif [ "UpperBounds" != "StateSpace" ] ; then
echo "The expected result is a vector of booleans"
echo BOOL_VECTOR
else
echo "no data necessary for post analysis"
fi
echo
if [ -f "UpperBounds.txt" ] ; then
echo "here is the order used to build the result vector(from text file)"
for x in $(grep Property UpperBounds.txt | cut -d ' ' -f 2 | sort -u) ; do
echo "FORMULA_NAME $x"
done
elif [ -f "UpperBounds.xml" ] ; then # for cunf (txt files deleted;-)
echo echo "here is the order used to build the result vector(from xml file)"
for x in $(grep '
echo "FORMULA_NAME $x"
done
fi
echo
echo "=== Now, execution of the tool begins"
echo
echo -n "BK_START "
date -u +%s%3N
echo
timeout -s 9 $BK_TIME_CONFINEMENT bash -c "/home/mcc/BenchKit/BenchKit_head.sh 2> STDERR ; echo ; echo -n \"BK_STOP \" ; date -u +%s%3N"
if [ $? -eq 137 ] ; then
echo
echo "BK_TIME_CONFINEMENT_REACHED"
fi
echo
echo "--------------------"
echo "content from stderr:"
echo
cat STDERR ;

