About the Execution of ITS-Tools for S_Angiogenesis-PT-01
| Execution Summary | |||||
| Max Memory Used (MB) |
Time wait (ms) | CPU Usage (ms) | I/O Wait (ms) | Computed Result | Execution Status |
| 227.470 | 2814.00 | 4084.00 | 68.80 | FFTFFFFFTFTFFFFF | normal |
Execution Chart
We display below the execution chart for this examination (boot time has been removed).
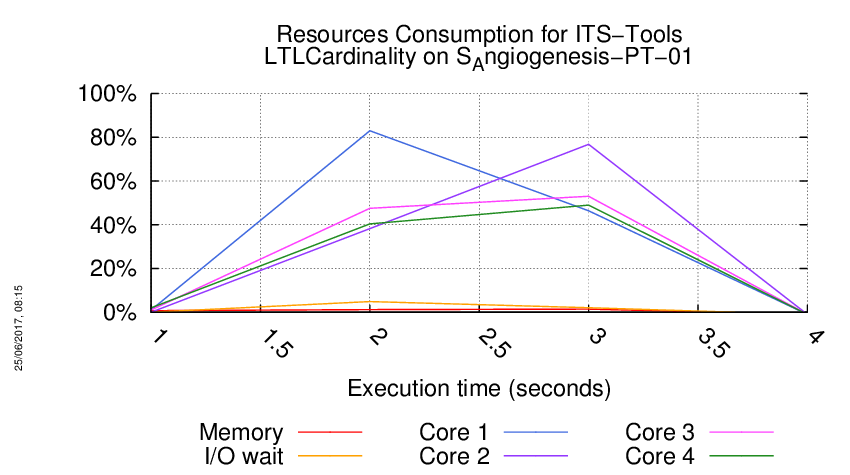
Trace from the execution
Waiting for the VM to be ready (probing ssh)
.......
=====================================================================
Generated by BenchKit 2-3254
Executing tool itstools
Input is S_Angiogenesis-PT-01, examination is LTLCardinality
Time confinement is 3600 seconds
Memory confinement is 16384 MBytes
Number of cores is 4
Run identifier is r090-csrt-149441076400113
=====================================================================
--------------------
content from stdout:
=== Data for post analysis generated by BenchKit (invocation template)
The expected result is a vector of booleans
BOOL_VECTOR
here is the order used to build the result vector(from text file)
FORMULA_NAME Angiogenesis-PT-01-LTLCardinality-0
FORMULA_NAME Angiogenesis-PT-01-LTLCardinality-1
FORMULA_NAME Angiogenesis-PT-01-LTLCardinality-10
FORMULA_NAME Angiogenesis-PT-01-LTLCardinality-11
FORMULA_NAME Angiogenesis-PT-01-LTLCardinality-12
FORMULA_NAME Angiogenesis-PT-01-LTLCardinality-13
FORMULA_NAME Angiogenesis-PT-01-LTLCardinality-14
FORMULA_NAME Angiogenesis-PT-01-LTLCardinality-15
FORMULA_NAME Angiogenesis-PT-01-LTLCardinality-2
FORMULA_NAME Angiogenesis-PT-01-LTLCardinality-3
FORMULA_NAME Angiogenesis-PT-01-LTLCardinality-4
FORMULA_NAME Angiogenesis-PT-01-LTLCardinality-5
FORMULA_NAME Angiogenesis-PT-01-LTLCardinality-6
FORMULA_NAME Angiogenesis-PT-01-LTLCardinality-7
FORMULA_NAME Angiogenesis-PT-01-LTLCardinality-8
FORMULA_NAME Angiogenesis-PT-01-LTLCardinality-9
=== Now, execution of the tool begins
BK_START 1496384355587
Using solver YICES2 to compute partial order matrices.
Built C files in :
/home/mcc/execution
its-ltl command run as :
/home/mcc/BenchKit/eclipse/plugins/fr.lip6.move.gal.itstools.binaries_1.0.0.201705302212/bin/its-ltl-linux64 --gc-threshold 2000000 -i /home/mcc/execution/LTLCardinality.pnml.gal -t CGAL -LTL /home/mcc/execution/LTLCardinality.ltl -c -stutter-deadlock
Read 16 LTL properties
Checking formula 0 : !((G("DAGE<=KdStarGStarP3kStarP2")))
Formula 0 simplified : !G"DAGE<=KdStarGStarP3kStarP2"
3 unique states visited
3 strongly connected components in search stack
3 transitions explored
3 items max in DFS search stack
7 ticks for the emptiness check
Model ,|S| ,Time ,Mem(kb) ,fin. SDD ,fin. DDD ,peak SDD ,peak DDD ,SDD Hom ,SDD cache peak ,DDD Hom ,DDD cachepeak ,SHom cache
STATS,0,0.09603,25788,1,0,91,9098,143,85,1408,9550,235
an accepting run exists (use option '-e' to print it)
Formula 0 is FALSE accepting run found.
FORMULA Angiogenesis-PT-01-LTLCardinality-0 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL
Checking formula 1 : !((X(X(F("DAGE>=1")))))
Formula 1 simplified : !XXF"DAGE>=1"
4 unique states visited
4 strongly connected components in search stack
4 transitions explored
4 items max in DFS search stack
0 ticks for the emptiness check
Model ,|S| ,Time ,Mem(kb) ,fin. SDD ,fin. DDD ,peak SDD ,peak DDD ,SDD Hom ,SDD cache peak ,DDD Hom ,DDD cachepeak ,SHom cache
STATS,0,0.097837,25984,1,0,93,9162,152,88,1409,9708,282
an accepting run exists (use option '-e' to print it)
Formula 1 is FALSE accepting run found.
FORMULA Angiogenesis-PT-01-LTLCardinality-1 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL
Checking formula 2 : !((F(F(("Akt>=3")U("KdStarGStarPgP3<=GStarPgP3")))))
Formula 2 simplified : !F("Akt>=3" U "KdStarGStarPgP3<=GStarPgP3")
1 unique states visited
0 strongly connected components in search stack
0 transitions explored
1 items max in DFS search stack
0 ticks for the emptiness check
Model ,|S| ,Time ,Mem(kb) ,fin. SDD ,fin. DDD ,peak SDD ,peak DDD ,SDD Hom ,SDD cache peak ,DDD Hom ,DDD cachepeak ,SHom cache
STATS,0,0.098262,26164,1,0,93,9162,161,88,1413,9710,286
no accepting run found
Formula 2 is TRUE no accepting run found.
FORMULA Angiogenesis-PT-01-LTLCardinality-2 TRUE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL
Checking formula 3 : !((G(F(("KdStarGStarPgStarP2>=3")U("KdStarGStarP3kP3>=3")))))
Formula 3 simplified : !GF("KdStarGStarPgStarP2>=3" U "KdStarGStarP3kP3>=3")
Presburger conditions satisfied. Using coverability to approximate state space in K-Induction.
3 unique states visited
3 strongly connected components in search stack
3 transitions explored
3 items max in DFS search stack
0 ticks for the emptiness check
Model ,|S| ,Time ,Mem(kb) ,fin. SDD ,fin. DDD ,peak SDD ,peak DDD ,SDD Hom ,SDD cache peak ,DDD Hom ,DDD cachepeak ,SHom cache
STATS,0,0.107863,26396,1,0,93,9309,170,88,1415,9931,294
an accepting run exists (use option '-e' to print it)
Formula 3 is FALSE accepting run found.
FORMULA Angiogenesis-PT-01-LTLCardinality-3 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL
Checking formula 4 : !((F(G(F(X("KdStarGP3<=Enz"))))))
Formula 4 simplified : !FGFX"KdStarGP3<=Enz"
2 unique states visited
0 strongly connected components in search stack
2 transitions explored
2 items max in DFS search stack
0 ticks for the emptiness check
Model ,|S| ,Time ,Mem(kb) ,fin. SDD ,fin. DDD ,peak SDD ,peak DDD ,SDD Hom ,SDD cache peak ,DDD Hom ,DDD cachepeak ,SHom cache
STATS,0,0.108714,26496,1,0,93,9309,179,88,1422,9943,298
no accepting run found
Formula 4 is TRUE no accepting run found.
FORMULA Angiogenesis-PT-01-LTLCardinality-4 TRUE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL
Checking formula 5 : !((F("KdStarGStarPgStar>=2")))
Formula 5 simplified : !F"KdStarGStarPgStar>=2"
2 unique states visited
2 strongly connected components in search stack
2 transitions explored
2 items max in DFS search stack
0 ticks for the emptiness check
Model ,|S| ,Time ,Mem(kb) ,fin. SDD ,fin. DDD ,peak SDD ,peak DDD ,SDD Hom ,SDD cache peak ,DDD Hom ,DDD cachepeak ,SHom cache
STATS,0,0.113549,26496,1,0,93,9654,188,102,1424,10874,385
an accepting run exists (use option '-e' to print it)
Formula 5 is FALSE accepting run found.
FORMULA Angiogenesis-PT-01-LTLCardinality-5 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL
Checking formula 6 : !((G(X(G("KdStarGStarP3k<=KdStarGStarPgStarP2")))))
Formula 6 simplified : !GXG"KdStarGStarP3k<=KdStarGStarPgStarP2"
4 unique states visited
4 strongly connected components in search stack
4 transitions explored
4 items max in DFS search stack
1 ticks for the emptiness check
Model ,|S| ,Time ,Mem(kb) ,fin. SDD ,fin. DDD ,peak SDD ,peak DDD ,SDD Hom ,SDD cache peak ,DDD Hom ,DDD cachepeak ,SHom cache
STATS,0,0.118767,26620,1,0,108,10075,194,118,1429,11674,438
an accepting run exists (use option '-e' to print it)
Formula 6 is FALSE accepting run found.
FORMULA Angiogenesis-PT-01-LTLCardinality-6 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL
Checking formula 7 : !((("DAG>=2")U(X("AktP3>=1"))))
Formula 7 simplified : !("DAG>=2" U X"AktP3>=1")
4 unique states visited
4 strongly connected components in search stack
4 transitions explored
4 items max in DFS search stack
0 ticks for the emptiness check
Model ,|S| ,Time ,Mem(kb) ,fin. SDD ,fin. DDD ,peak SDD ,peak DDD ,SDD Hom ,SDD cache peak ,DDD Hom ,DDD cachepeak ,SHom cache
STATS,0,0.12083,26840,1,0,108,10138,201,118,1432,11831,446
an accepting run exists (use option '-e' to print it)
Formula 7 is FALSE accepting run found.
FORMULA Angiogenesis-PT-01-LTLCardinality-7 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL
Checking formula 8 : !((X(G(X(G("KdStarGP3>=3"))))))
Formula 8 simplified : !XGXG"KdStarGP3>=3"
5 unique states visited
5 strongly connected components in search stack
5 transitions explored
5 items max in DFS search stack
0 ticks for the emptiness check
Model ,|S| ,Time ,Mem(kb) ,fin. SDD ,fin. DDD ,peak SDD ,peak DDD ,SDD Hom ,SDD cache peak ,DDD Hom ,DDD cachepeak ,SHom cache
STATS,0,0.12426,26904,1,0,108,10225,207,118,1434,12006,453
an accepting run exists (use option '-e' to print it)
Formula 8 is FALSE accepting run found.
FORMULA Angiogenesis-PT-01-LTLCardinality-8 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL
Checking formula 9 : !((F(G(X(F("KdStarG>=2"))))))
Formula 9 simplified : !FGXF"KdStarG>=2"
3 unique states visited
3 strongly connected components in search stack
4 transitions explored
3 items max in DFS search stack
0 ticks for the emptiness check
Model ,|S| ,Time ,Mem(kb) ,fin. SDD ,fin. DDD ,peak SDD ,peak DDD ,SDD Hom ,SDD cache peak ,DDD Hom ,DDD cachepeak ,SHom cache
STATS,0,0.125414,26968,1,0,108,10225,216,118,1436,12015,459
an accepting run exists (use option '-e' to print it)
Formula 9 is FALSE accepting run found.
FORMULA Angiogenesis-PT-01-LTLCardinality-9 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL
Checking formula 10 : !(("KdStarGStarP3kStarP3P2<=Pten"))
Formula 10 simplified : !"KdStarGStarP3kStarP3P2<=Pten"
1 unique states visited
0 strongly connected components in search stack
0 transitions explored
1 items max in DFS search stack
0 ticks for the emptiness check
Model ,|S| ,Time ,Mem(kb) ,fin. SDD ,fin. DDD ,peak SDD ,peak DDD ,SDD Hom ,SDD cache peak ,DDD Hom ,DDD cachepeak ,SHom cache
// Phase 1: matrix 64 rows 39 cols
invariant : 1'GStarP3kP3 + 1'KdStarGStarP3k + 1'KdStarGStarP3kP3 + 1'KdStarGStarP3kStar + 1'KdStarGStarP3kStarP2 + 1'KdStarGStarP3kStarP3 + 1'KdStarGStarP3kStarP3P2 + 1'P3k= 1
invariant : 1'GStarPgP3 + -1'KdStar + -1'KdStarG + -1'KdStarGP3 + -1'KdStarGStar + -1'KdStarGStarP3 + -1'KdStarGStarP3k + -1'KdStarGStarP3kP3 + -1'KdStarGStarP3kStar + -1'KdStarGStarP3kStarP2 + -1'KdStarGStarP3kStarP3 + -1'KdStarGStarP3kStarP3P2 + 1'Pg= 0
invariant : -1'AktP3 + -1'DAG + -1'DAGE + 1'Gab1 + 1'KdStar + 2'KdStarG + 1'KdStarGP3 + 2'KdStarGStar + 1'KdStarGStarP3 + 2'KdStarGStarP3k + 1'KdStarGStarP3kP3 + 2'KdStarGStarP3kStar + 1'KdStarGStarP3kStarP2 + 1'KdStarGStarP3kStarP3 + 2'KdStarGStarPg + 1'KdStarGStarPgP3 + 2'KdStarGStarPgStar + 1'KdStarGStarPgStarP2 + 1'KdStarGStarPgStarP3 + 1'KdStarPg + 1'KdStarPgStar + -1'Pip2 + -1'Pip3 + 2'Pten + 1'PtP2 + 1'PtP3= 3
invariant : 1'Akt + 1'AktP3 + 1'AktStar= 1
invariant : 1'DAGE + 1'Enz= 1
invariant : -1'Gab1 + -1'GP3 + -1'GStarP3 + -1'GStarP3kP3 + -1'GStarPgP3 + 1'KdStar + 1'KdStarPg + 1'KdStarPgStar + 1'KdStarPgStarP2= 0
invariant : 1'AktP3 + 1'DAG + 1'DAGE + -1'Gab1 + -1'KdStar + -2'KdStarG + -1'KdStarGP3 + -2'KdStarGStar + -1'KdStarGStarP3 + -2'KdStarGStarP3k + -1'KdStarGStarP3kP3 + -2'KdStarGStarP3kStar + -1'KdStarGStarP3kStarP2 + -1'KdStarGStarP3kStarP3 + -2'KdStarGStarPg + -1'KdStarGStarPgP3 + -2'KdStarGStarPgStar + -1'KdStarGStarPgStarP2 + -1'KdStarGStarPgStarP3 + -1'KdStarPg + -1'KdStarPgStar + 1'Pip2 + 1'Pip3 + -1'Pten + 1'PtP3P2= -2
invariant : 1'Gab1 + 1'GP3 + 1'GStarP3 + 1'GStarP3kP3 + 1'GStarPgP3 + 1'KdStarG + 1'KdStarGP3 + 1'KdStarGStar + 1'KdStarGStarP3 + 1'KdStarGStarP3k + 1'KdStarGStarP3kP3 + 1'KdStarGStarP3kStar + 1'KdStarGStarP3kStarP2 + 1'KdStarGStarP3kStarP3 + 1'KdStarGStarP3kStarP3P2 + 1'KdStarGStarPg + 1'KdStarGStarPgP3 + 1'KdStarGStarPgStar + 1'KdStarGStarPgStarP2 + 1'KdStarGStarPgStarP3 + 1'KdStarGStarPgStarP3P2= 1
STATS,0,0.125866,26968,1,0,108,10225,219,118,1441,12016,461
no accepting run found
Formula 10 is TRUE no accepting run found.
FORMULA Angiogenesis-PT-01-LTLCardinality-10 TRUE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL
Checking formula 11 : !((G("KdStarG>=1")))
Formula 11 simplified : !G"KdStarG>=1"
3 unique states visited
3 strongly connected components in search stack
3 transitions explored
3 items max in DFS search stack
0 ticks for the emptiness check
Model ,|S| ,Time ,Mem(kb) ,fin. SDD ,fin. DDD ,peak SDD ,peak DDD ,SDD Hom ,SDD cache peak ,DDD Hom ,DDD cachepeak ,SHom cache
STATS,0,0.126771,26976,1,0,108,10226,225,118,1442,12017,468
an accepting run exists (use option '-e' to print it)
Formula 11 is FALSE accepting run found.
FORMULA Angiogenesis-PT-01-LTLCardinality-11 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL
Checking formula 12 : !((G("KdStarGP3>=1")))
Formula 12 simplified : !G"KdStarGP3>=1"
3 unique states visited
3 strongly connected components in search stack
3 transitions explored
3 items max in DFS search stack
0 ticks for the emptiness check
Model ,|S| ,Time ,Mem(kb) ,fin. SDD ,fin. DDD ,peak SDD ,peak DDD ,SDD Hom ,SDD cache peak ,DDD Hom ,DDD cachepeak ,SHom cache
STATS,0,0.12742,26980,1,0,108,10226,231,118,1443,12017,473
an accepting run exists (use option '-e' to print it)
Formula 12 is FALSE accepting run found.
FORMULA Angiogenesis-PT-01-LTLCardinality-12 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL
Checking formula 13 : !((F(G(G(G("KdStarGStarPgP3>=1"))))))
Formula 13 simplified : !FG"KdStarGStarPgP3>=1"
2 unique states visited
2 strongly connected components in search stack
2 transitions explored
2 items max in DFS search stack
1 ticks for the emptiness check
Model ,|S| ,Time ,Mem(kb) ,fin. SDD ,fin. DDD ,peak SDD ,peak DDD ,SDD Hom ,SDD cache peak ,DDD Hom ,DDD cachepeak ,SHom cache
STATS,0,0.138595,26992,1,0,136,10633,244,154,1444,13350,599
an accepting run exists (use option '-e' to print it)
Formula 13 is FALSE accepting run found.
FORMULA Angiogenesis-PT-01-LTLCardinality-13 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL
Checking formula 14 : !((G(F("KdStarGStarP3>=1"))))
Formula 14 simplified : !GF"KdStarGStarP3>=1"
3 unique states visited
3 strongly connected components in search stack
4 transitions explored
3 items max in DFS search stack
0 ticks for the emptiness check
Model ,|S| ,Time ,Mem(kb) ,fin. SDD ,fin. DDD ,peak SDD ,peak DDD ,SDD Hom ,SDD cache peak ,DDD Hom ,DDD cachepeak ,SHom cache
STATS,0,0.139869,27072,1,0,137,10639,253,155,1445,13394,608
an accepting run exists (use option '-e' to print it)
Formula 14 is FALSE accepting run found.
FORMULA Angiogenesis-PT-01-LTLCardinality-14 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL
Checking formula 15 : !((X(F("KdStarGStarPgStar>=1"))))
Formula 15 simplified : !XF"KdStarGStarPgStar>=1"
3 unique states visited
3 strongly connected components in search stack
3 transitions explored
3 items max in DFS search stack
1 ticks for the emptiness check
Model ,|S| ,Time ,Mem(kb) ,fin. SDD ,fin. DDD ,peak SDD ,peak DDD ,SDD Hom ,SDD cache peak ,DDD Hom ,DDD cachepeak ,SHom cache
STATS,0,0.146028,27164,1,0,158,11143,262,180,1446,14597,674
an accepting run exists (use option '-e' to print it)
Formula 15 is FALSE accepting run found.
FORMULA Angiogenesis-PT-01-LTLCardinality-15 FALSE TECHNIQUES DECISION_DIAGRAMS TOPOLOGICAL
Exit code :0
BK_STOP 1496384358401
--------------------
content from stderr:
+ export BINDIR=/home/mcc/BenchKit/
+ BINDIR=/home/mcc/BenchKit/
++ pwd
+ export MODEL=/home/mcc/execution
+ MODEL=/home/mcc/execution
+ [[ LTLCardinality = StateSpace ]]
+ /home/mcc/BenchKit//runeclipse.sh /home/mcc/execution LTLCardinality -its -ltsminpath /home/mcc/BenchKit//lts_install_dir/ -smt
+ ulimit -s 65536
+ java -Dosgi.requiredJavaVersion=1.6 -Xss8m -Xms40m -Xmx8192m -Declipse.pde.launch=true -Dfile.encoding=UTF-8 -classpath /home/mcc/BenchKit//eclipse/plugins/org.eclipse.equinox.launcher_1.3.201.v20161025-1711.jar org.eclipse.equinox.launcher.Main -application fr.lip6.move.gal.application.pnmcc -data /home/mcc/BenchKit//workspace -os linux -ws gtk -arch x86_64 -nl en_US -consoleLog -pnfolder /home/mcc/execution -examination LTLCardinality -yices2path /home/mcc/BenchKit//yices/bin/yices -its -ltsminpath /home/mcc/BenchKit//lts_install_dir/ -smt
Jun 02, 2017 6:19:17 AM fr.lip6.move.gal.application.MccTranslator transformPNML
INFO: Parsing pnml file : /home/mcc/execution/model.pnml
Jun 02, 2017 6:19:17 AM fr.lip6.move.gal.nupn.PTNetReader loadFromXML
INFO: Load time of PNML (sax parser for PT used): 45 ms
Jun 02, 2017 6:19:17 AM fr.lip6.move.gal.pnml.togal.PTGALTransformer handlePage
INFO: Transformed 39 places.
Jun 02, 2017 6:19:17 AM fr.lip6.move.gal.pnml.togal.PTGALTransformer handlePage
INFO: Transformed 64 transitions.
Jun 02, 2017 6:19:17 AM fr.lip6.move.gal.instantiate.GALRewriter flatten
INFO: Flatten gal took : 78 ms
Jun 02, 2017 6:19:17 AM fr.lip6.move.serialization.SerializationUtil systemToFile
INFO: Time to serialize gal into /home/mcc/execution/LTLCardinality.pnml.gal : 23 ms
Jun 02, 2017 6:19:17 AM fr.lip6.move.serialization.SerializationUtil serializePropertiesForITSLTLTools
INFO: Time to serialize properties into /home/mcc/execution/LTLCardinality.ltl : 1 ms
Jun 02, 2017 6:19:18 AM fr.lip6.move.gal.gal2smt.bmc.KInductionSolver computeAndDeclareInvariants
INFO: Computed 8 place invariants in 34 ms
Jun 02, 2017 6:19:18 AM fr.lip6.move.gal.itstools.Runner runTool
INFO: Standard error output from running tool CommandLine [args=[/home/mcc/BenchKit/eclipse/plugins/fr.lip6.move.gal.itstools.binaries_1.0.0.201705302212/bin/its-ltl-linux64, --gc-threshold, 2000000, -i, /home/mcc/execution/LTLCardinality.pnml.gal, -t, CGAL, -LTL, /home/mcc/execution/LTLCardinality.ltl, -c, -stutter-deadlock], workingDir=/home/mcc/execution]
Reverse transition relation is NOT exact ! Due to transitions k2, k55, Intersection with reachable at each step enabled. (destroyed/reverse/intersect/total) :14/48/2/64
Computing Next relation with stutter on 4 deadlock states
Computing Next relation with stutter on 4 deadlock states
Computing Next relation with stutter on 4 deadlock states
Computing Next relation with stutter on 4 deadlock states
Computing Next relation with stutter on 4 deadlock states
Computing Next relation with stutter on 4 deadlock states
Computing Next relation with stutter on 4 deadlock states
Computing Next relation with stutter on 4 deadlock states
Computing Next relation with stutter on 4 deadlock states
Computing Next relation with stutter on 4 deadlock states
Computing Next relation with stutter on 4 deadlock states
Computing Next relation with stutter on 4 deadlock states
Computing Next relation with stutter on 4 deadlock states
Computing Next relation with stutter on 4 deadlock states
Computing Next relation with stutter on 4 deadlock states
Computing Next relation with stutter on 4 deadlock states
Sequence of Actions to be Executed by the VM
This is useful if one wants to reexecute the tool in the VM from the submitted image disk.
set -x
# this is for BenchKit: configuration of major elements for the test
export BK_INPUT="S_Angiogenesis-PT-01"
export BK_EXAMINATION="LTLCardinality"
export BK_TOOL="itstools"
export BK_RESULT_DIR="/tmp/BK_RESULTS/OUTPUTS"
export BK_TIME_CONFINEMENT="3600"
export BK_MEMORY_CONFINEMENT="16384"
# this is specific to your benchmark or test
export BIN_DIR="$HOME/BenchKit/bin"
# remove the execution directoty if it exists (to avoid increse of .vmdk images)
if [ -d execution ] ; then
rm -rf execution
fi
tar xzf /home/mcc/BenchKit/INPUTS/S_Angiogenesis-PT-01.tgz
mv S_Angiogenesis-PT-01 execution
# this is for BenchKit: explicit launching of the test
cd execution
echo "====================================================================="
echo " Generated by BenchKit 2-3254"
echo " Executing tool itstools"
echo " Input is S_Angiogenesis-PT-01, examination is LTLCardinality"
echo " Time confinement is $BK_TIME_CONFINEMENT seconds"
echo " Memory confinement is 16384 MBytes"
echo " Number of cores is 4"
echo " Run identifier is r090-csrt-149441076400113"
echo "====================================================================="
echo
echo "--------------------"
echo "content from stdout:"
echo
echo "=== Data for post analysis generated by BenchKit (invocation template)"
echo
if [ "LTLCardinality" = "UpperBounds" ] ; then
echo "The expected result is a vector of positive values"
echo NUM_VECTOR
elif [ "LTLCardinality" != "StateSpace" ] ; then
echo "The expected result is a vector of booleans"
echo BOOL_VECTOR
else
echo "no data necessary for post analysis"
fi
echo
if [ -f "LTLCardinality.txt" ] ; then
echo "here is the order used to build the result vector(from text file)"
for x in $(grep Property LTLCardinality.txt | cut -d ' ' -f 2 | sort -u) ; do
echo "FORMULA_NAME $x"
done
elif [ -f "LTLCardinality.xml" ] ; then # for cunf (txt files deleted;-)
echo echo "here is the order used to build the result vector(from xml file)"
for x in $(grep '
echo "FORMULA_NAME $x"
done
fi
echo
echo "=== Now, execution of the tool begins"
echo
echo -n "BK_START "
date -u +%s%3N
echo
timeout -s 9 $BK_TIME_CONFINEMENT bash -c "/home/mcc/BenchKit/BenchKit_head.sh 2> STDERR ; echo ; echo -n \"BK_STOP \" ; date -u +%s%3N"
if [ $? -eq 137 ] ; then
echo
echo "BK_TIME_CONFINEMENT_REACHED"
fi
echo
echo "--------------------"
echo "content from stderr:"
echo
cat STDERR ;

