About the Execution of MARCIE for DNAwalker-PT-02track12Block2
| Execution Summary | |||||
| Max Memory Used (MB) |
Time wait (ms) | CPU Usage (ms) | I/O Wait (ms) | Computed Result | Execution Status |
| 7483.090 | 5895.00 | 4990.00 | 30.00 | FFTTTTFFFFFTTTTF | normal |
Execution Chart
We display below the execution chart for this examination (boot time has been removed).
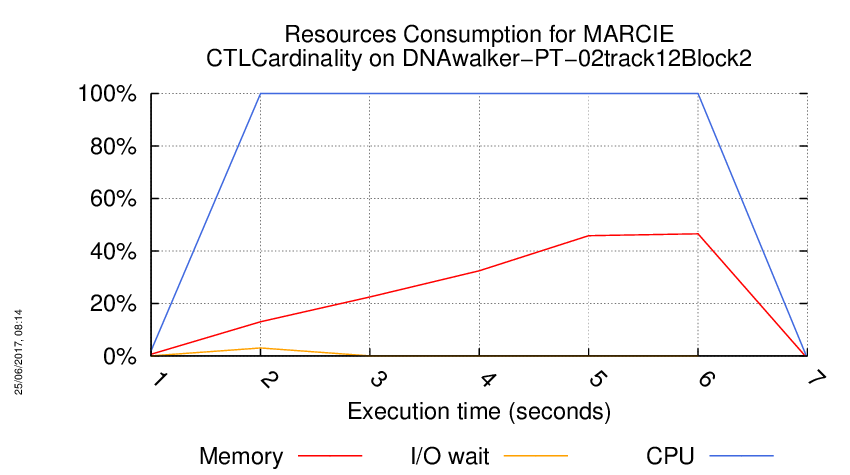
Trace from the execution
Waiting for the VM to be ready (probing ssh)
......
=====================================================================
Generated by BenchKit 2-3254
Executing tool marcie
Input is DNAwalker-PT-02track12Block2, examination is CTLCardinality
Time confinement is 3600 seconds
Memory confinement is 16384 MBytes
Number of cores is 1
Run identifier is r021-blw3-149440254700012
=====================================================================
--------------------
content from stdout:
=== Data for post analysis generated by BenchKit (invocation template)
The expected result is a vector of booleans
BOOL_VECTOR
here is the order used to build the result vector(from text file)
FORMULA_NAME DNAwalker-PT-02track12Block2-CTLCardinality-0
FORMULA_NAME DNAwalker-PT-02track12Block2-CTLCardinality-1
FORMULA_NAME DNAwalker-PT-02track12Block2-CTLCardinality-10
FORMULA_NAME DNAwalker-PT-02track12Block2-CTLCardinality-11
FORMULA_NAME DNAwalker-PT-02track12Block2-CTLCardinality-12
FORMULA_NAME DNAwalker-PT-02track12Block2-CTLCardinality-13
FORMULA_NAME DNAwalker-PT-02track12Block2-CTLCardinality-14
FORMULA_NAME DNAwalker-PT-02track12Block2-CTLCardinality-15
FORMULA_NAME DNAwalker-PT-02track12Block2-CTLCardinality-2
FORMULA_NAME DNAwalker-PT-02track12Block2-CTLCardinality-3
FORMULA_NAME DNAwalker-PT-02track12Block2-CTLCardinality-4
FORMULA_NAME DNAwalker-PT-02track12Block2-CTLCardinality-5
FORMULA_NAME DNAwalker-PT-02track12Block2-CTLCardinality-6
FORMULA_NAME DNAwalker-PT-02track12Block2-CTLCardinality-7
FORMULA_NAME DNAwalker-PT-02track12Block2-CTLCardinality-8
FORMULA_NAME DNAwalker-PT-02track12Block2-CTLCardinality-9
=== Now, execution of the tool begins
BK_START 1494448532318
timeout --kill-after=10s --signal=SIGINT 1m for testing only
Marcie rev. 8852M (built: crohr on 2017-05-03)
A model checker for Generalized Stochastic Petri nets
authors: Alex Tovchigrechko (IDD package and CTL model checking)
Martin Schwarick (Symbolic numerical analysis and CSL model checking)
Christian Rohr (Simulative and approximative numerical model checking)
marcie@informatik.tu-cottbus.de
called as: marcie --net-file=model.pnml --mcc-file=CTLCardinality.xml --memory=6
parse successfull
net created successfully
Net: DNAwalker_PT_02track12Block2
(NrP: 14 NrTr: 84 NrArc: 244)
parse formulas
formulas created successfully
place and transition orderings generation:0m 0.000sec
net check time: 0m 0.000sec
init dd package: 0m 1.038sec
parse successfull
net created successfully
Net: DNAwalker_PT_02track12Block2
(NrP: 14 NrTr: 84 NrArc: 244)
parse formulas
formulas created successfully
place and transition orderings generation:0m 0.000sec
net check time: 0m 0.000sec
init dd package: 0m 3.269sec
RS generation: 0m 0.022sec
-> reachability set: #nodes 178 (1.8e+02) #states 5,459 (3)
starting MCC model checker
--------------------------
checking: AG [~ [EF [2<=B5]]]
normalized: ~ [E [true U E [true U 2<=B5]]]
abstracting: (2<=B5)
states: 0
-> the formula is TRUE
FORMULA DNAwalker-PT-02track12Block2-CTLCardinality-7 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.000sec
checking: EF [AG [~ [1<=A6]]]
normalized: E [true U ~ [E [true U 1<=A6]]]
abstracting: (1<=A6)
states: 1,925 (3)
-> the formula is TRUE
FORMULA DNAwalker-PT-02track12Block2-CTLCardinality-8 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.024sec
checking: AF [AG [~ [2<=B5]]]
normalized: ~ [EG [E [true U 2<=B5]]]
abstracting: (2<=B5)
states: 0
.
EG iterations: 1
-> the formula is TRUE
FORMULA DNAwalker-PT-02track12Block2-CTLCardinality-11 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.002sec
checking: EG [EF [3<=A12]]
normalized: EG [E [true U 3<=A12]]
abstracting: (3<=A12)
states: 0
.
EG iterations: 1
-> the formula is FALSE
FORMULA DNAwalker-PT-02track12Block2-CTLCardinality-15 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.000sec
checking: EX [~ [EG [A12<=A11]]]
normalized: EX [~ [EG [A12<=A11]]]
abstracting: (A12<=A11)
states: 2,728 (3)
.......
EG iterations: 7
.-> the formula is FALSE
FORMULA DNAwalker-PT-02track12Block2-CTLCardinality-3 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.023sec
checking: EF [EG [[1<=A2 | A6<=B6]]]
normalized: E [true U EG [[1<=A2 | A6<=B6]]]
abstracting: (A6<=B6)
states: 3,534 (3)
abstracting: (1<=A2)
states: 2,805 (3)
...
EG iterations: 3
-> the formula is TRUE
FORMULA DNAwalker-PT-02track12Block2-CTLCardinality-5 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.003sec
checking: [~ [2<=A7] & EG [AX [B6<=A10]]]
normalized: [EG [~ [EX [~ [B6<=A10]]]] & ~ [2<=A7]]
abstracting: (2<=A7)
states: 462
abstracting: (B6<=A10)
states: 4,705 (3)
..
EG iterations: 1
-> the formula is TRUE
FORMULA DNAwalker-PT-02track12Block2-CTLCardinality-10 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.008sec
checking: AF [[2<=A10 & ~ [[A1<=A12 | 1<=A5]]]]
normalized: ~ [EG [~ [[2<=A10 & ~ [[A1<=A12 | 1<=A5]]]]]]
abstracting: (1<=A5)
states: 1,850 (3)
abstracting: (A1<=A12)
states: 5,450 (3)
abstracting: (2<=A10)
states: 471
EG iterations: 0
-> the formula is FALSE
FORMULA DNAwalker-PT-02track12Block2-CTLCardinality-0 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.000sec
checking: [AF [A3<=A10] & ~ [AF [[1<=A6 & A10<=B6]]]]
normalized: [EG [~ [[1<=A6 & A10<=B6]]] & ~ [EG [~ [A3<=A10]]]]
abstracting: (A3<=A10)
states: 3,970 (3)
....
EG iterations: 4
abstracting: (A10<=B6)
states: 3,317 (3)
abstracting: (1<=A6)
states: 1,925 (3)
...
EG iterations: 3
-> the formula is TRUE
FORMULA DNAwalker-PT-02track12Block2-CTLCardinality-13 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.015sec
checking: [~ [AX [~ [B5<=A2]]] & E [A8<=B5 U [2<=B5 | 3<=A11]]]
normalized: [E [A8<=B5 U [2<=B5 | 3<=A11]] & EX [B5<=A2]]
abstracting: (B5<=A2)
states: 4,703 (3)
.abstracting: (3<=A11)
states: 0
abstracting: (2<=B5)
states: 0
abstracting: (A8<=B5)
states: 1,335 (3)
-> the formula is FALSE
FORMULA DNAwalker-PT-02track12Block2-CTLCardinality-1 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.001sec
checking: EG [[AF [B5<=B5] | [~ [A7<=A5] & [3<=A1 | 3<=A6]]]]
normalized: EG [[[[3<=A1 | 3<=A6] & ~ [A7<=A5]] | ~ [EG [~ [B5<=B5]]]]]
abstracting: (B5<=B5)
states: 5,459 (3)
.
EG iterations: 1
abstracting: (A7<=A5)
states: 3,242 (3)
abstracting: (3<=A6)
states: 0
abstracting: (3<=A1)
states: 0
EG iterations: 0
-> the formula is TRUE
FORMULA DNAwalker-PT-02track12Block2-CTLCardinality-6 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.000sec
checking: [[A3<=A9 | E [2<=A12 U 1<=A6]] & A [~ [3<=A12] U 3<=A1]]
normalized: [[~ [EG [~ [3<=A1]]] & ~ [E [~ [3<=A1] U [~ [3<=A1] & 3<=A12]]]] & [A3<=A9 | E [2<=A12 U 1<=A6]]]
abstracting: (1<=A6)
states: 1,925 (3)
abstracting: (2<=A12)
states: 847
abstracting: (A3<=A9)
states: 3,759 (3)
abstracting: (3<=A12)
states: 0
abstracting: (3<=A1)
states: 0
abstracting: (3<=A1)
states: 0
abstracting: (3<=A1)
states: 0
EG iterations: 0
-> the formula is FALSE
FORMULA DNAwalker-PT-02track12Block2-CTLCardinality-9 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.003sec
checking: [~ [[AX [3<=A9] | [~ [3<=B6] | [2<=A4 | 2<=A12]]]] | EG [EF [A1<=A6]]]
normalized: [EG [E [true U A1<=A6]] | ~ [[[[2<=A4 | 2<=A12] | ~ [3<=B6]] | ~ [EX [~ [3<=A9]]]]]]
abstracting: (3<=A9)
states: 0
.abstracting: (3<=B6)
states: 0
abstracting: (2<=A12)
states: 847
abstracting: (2<=A4)
states: 463
abstracting: (A1<=A6)
states: 5,450 (3)
EG iterations: 0
-> the formula is TRUE
FORMULA DNAwalker-PT-02track12Block2-CTLCardinality-12 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.000sec
checking: [AF [EG [2<=B5]] & [EX [[A2<=A11 & 1<=A5]] | [EX [3<=B6] | AF [A12<=B6]]]]
normalized: [[[~ [EG [~ [A12<=B6]]] | EX [3<=B6]] | EX [[A2<=A11 & 1<=A5]]] & ~ [EG [~ [EG [2<=B5]]]]]
abstracting: (2<=B5)
states: 0
.
EG iterations: 1
EG iterations: 0
abstracting: (1<=A5)
states: 1,850 (3)
abstracting: (A2<=A11)
states: 4,014 (3)
.abstracting: (3<=B6)
states: 0
.abstracting: (A12<=B6)
states: 1,341 (3)
.
EG iterations: 1
-> the formula is FALSE
FORMULA DNAwalker-PT-02track12Block2-CTLCardinality-4 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.007sec
checking: [E [~ [B6<=A5] U 3<=B6] & [[AF [3<=A11] & [[2<=A5 & 3<=A5] | [A5<=B6 | 1<=A3]]] & B6<=A7]]
normalized: [[[[[A5<=B6 | 1<=A3] | [2<=A5 & 3<=A5]] & ~ [EG [~ [3<=A11]]]] & B6<=A7] & E [~ [B6<=A5] U 3<=B6]]
abstracting: (3<=B6)
states: 0
abstracting: (B6<=A5)
states: 4,384 (3)
abstracting: (B6<=A7)
states: 4,806 (3)
abstracting: (3<=A11)
states: 0
EG iterations: 0
abstracting: (3<=A5)
states: 0
abstracting: (2<=A5)
states: 323
abstracting: (1<=A3)
states: 2,594 (3)
abstracting: (A5<=B6)
states: 4,041 (3)
-> the formula is FALSE
FORMULA DNAwalker-PT-02track12Block2-CTLCardinality-2 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.001sec
checking: AF [[[[A7<=A5 & 3<=A9] & [A9<=A3 & 2<=A4]] & [[B5<=A2 | 3<=A4] | [A3<=A11 & 1<=A8]]]]
normalized: ~ [EG [~ [[[[A3<=A11 & 1<=A8] | [B5<=A2 | 3<=A4]] & [[A9<=A3 & 2<=A4] & [A7<=A5 & 3<=A9]]]]]]
abstracting: (3<=A9)
states: 0
abstracting: (A7<=A5)
states: 3,242 (3)
abstracting: (2<=A4)
states: 463
abstracting: (A9<=A3)
states: 3,821 (3)
abstracting: (3<=A4)
states: 0
abstracting: (B5<=A2)
states: 4,703 (3)
abstracting: (1<=A8)
states: 5,459 (3)
abstracting: (A3<=A11)
states: 4,139 (3)
EG iterations: 0
-> the formula is FALSE
FORMULA DNAwalker-PT-02track12Block2-CTLCardinality-14 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.000sec
totally nodes used: 44392 (4.4e+04)
number of garbage collections: 0
fire ops cache: hits/miss/sum: 94499 154035 248534
used/not used/entry size/cache size: 194427 66914437 16 1024MB
basic ops cache: hits/miss/sum: 18850 20141 38991
used/not used/entry size/cache size: 36714 16740502 12 192MB
unary ops cache: hits/miss/sum: 0 0 0
used/not used/entry size/cache size: 0 16777216 8 128MB
abstract ops cache: hits/miss/sum: 0 0 0
used/not used/entry size/cache size: 0 16777216 12 192MB
state nr cache: hits/miss/sum: 1501 1705 3206
used/not used/entry size/cache size: 1705 8386903 32 256MB
max state cache: hits/miss/sum: 0 0 0
used/not used/entry size/cache size: 0 8388608 32 256MB
uniqueHash elements/entry size/size: 67108864 4 256MB
0 67068367
1 37720
2 2055
3 494
4 128
5 69
6 17
7 7
8 3
9 2
>= 10 2
Total processing time: 0m 5.809sec
BK_STOP 1494448538213
--------------------
content from stderr:
check for maximal unmarked siphon
ok
check for constant places
ok
check if there are places and transitions
ok
check if there are transitions without pre-places
ok
check if at least one transition is enabled in m0
ok
check if there are transitions that can never fire
ok
ptnet_zbdd.cc:255: Boundedness exception: net is not 1-bounded!
check for maximal unmarked siphon
ok
check for constant places
ok
check if there are places and transitions
ok
check if there are transitions without pre-places
ok
check if at least one transition is enabled in m0
ok
check if there are transitions that can never fire
ok
.........10 178........................
Sequence of Actions to be Executed by the VM
This is useful if one wants to reexecute the tool in the VM from the submitted image disk.
set -x
# this is for BenchKit: configuration of major elements for the test
export BK_INPUT="DNAwalker-PT-02track12Block2"
export BK_EXAMINATION="CTLCardinality"
export BK_TOOL="marcie"
export BK_RESULT_DIR="/tmp/BK_RESULTS/OUTPUTS"
export BK_TIME_CONFINEMENT="3600"
export BK_MEMORY_CONFINEMENT="16384"
# this is specific to your benchmark or test
export BIN_DIR="$HOME/BenchKit/bin"
# remove the execution directoty if it exists (to avoid increse of .vmdk images)
if [ -d execution ] ; then
rm -rf execution
fi
tar xzf /home/mcc/BenchKit/INPUTS/DNAwalker-PT-02track12Block2.tgz
mv DNAwalker-PT-02track12Block2 execution
# this is for BenchKit: explicit launching of the test
cd execution
echo "====================================================================="
echo " Generated by BenchKit 2-3254"
echo " Executing tool marcie"
echo " Input is DNAwalker-PT-02track12Block2, examination is CTLCardinality"
echo " Time confinement is $BK_TIME_CONFINEMENT seconds"
echo " Memory confinement is 16384 MBytes"
echo " Number of cores is 1"
echo " Run identifier is r021-blw3-149440254700012"
echo "====================================================================="
echo
echo "--------------------"
echo "content from stdout:"
echo
echo "=== Data for post analysis generated by BenchKit (invocation template)"
echo
if [ "CTLCardinality" = "UpperBounds" ] ; then
echo "The expected result is a vector of positive values"
echo NUM_VECTOR
elif [ "CTLCardinality" != "StateSpace" ] ; then
echo "The expected result is a vector of booleans"
echo BOOL_VECTOR
else
echo "no data necessary for post analysis"
fi
echo
if [ -f "CTLCardinality.txt" ] ; then
echo "here is the order used to build the result vector(from text file)"
for x in $(grep Property CTLCardinality.txt | cut -d ' ' -f 2 | sort -u) ; do
echo "FORMULA_NAME $x"
done
elif [ -f "CTLCardinality.xml" ] ; then # for cunf (txt files deleted;-)
echo echo "here is the order used to build the result vector(from xml file)"
for x in $(grep '
echo "FORMULA_NAME $x"
done
fi
echo
echo "=== Now, execution of the tool begins"
echo
echo -n "BK_START "
date -u +%s%3N
echo
timeout -s 9 $BK_TIME_CONFINEMENT bash -c "/home/mcc/BenchKit/BenchKit_head.sh 2> STDERR ; echo ; echo -n \"BK_STOP \" ; date -u +%s%3N"
if [ $? -eq 137 ] ; then
echo
echo "BK_TIME_CONFINEMENT_REACHED"
fi
echo
echo "--------------------"
echo "content from stderr:"
echo
cat STDERR ;

