About the Execution of LTSMin for Angiogenesis-PT-10
| Execution Summary | |||||
| Max Memory Used (MB) |
Time wait (ms) | CPU Usage (ms) | I/O Wait (ms) | Computed Result | Execution Status |
| 15528.760 | 3572534.00 | 14281910.00 | 10.10 | TT?????????????? | normal |
Execution Chart
We display below the execution chart for this examination (boot time has been removed).
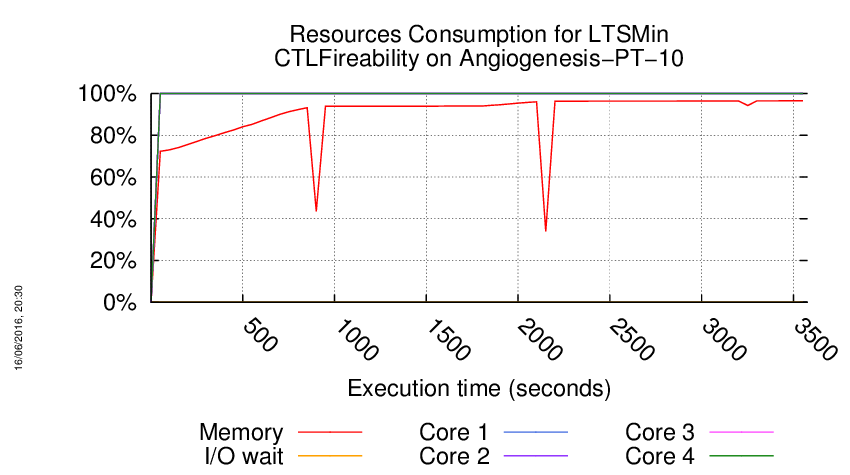
Trace from the execution
Waiting for the VM to be ready (probing ssh)
............
=====================================================================
Generated by BenchKit 2-2979
Executing tool ltsmin
Input is Angiogenesis-PT-10, examination is CTLFireability
Time confinement is 3600 seconds
Memory confinement is 16384 MBytes
Number of cores is 4
Run identifier is r003kn-ebro-146347663700031
=====================================================================
--------------------
content from stdout:
=== Data for post analysis generated by BenchKit (invocation template)
The expected result is a vector of booleans
BOOL_VECTOR
here is the order used to build the result vector(from text file)
FORMULA_NAME Angiogenesis-PT-10-CTLFireability-0
FORMULA_NAME Angiogenesis-PT-10-CTLFireability-1
FORMULA_NAME Angiogenesis-PT-10-CTLFireability-10
FORMULA_NAME Angiogenesis-PT-10-CTLFireability-11
FORMULA_NAME Angiogenesis-PT-10-CTLFireability-12
FORMULA_NAME Angiogenesis-PT-10-CTLFireability-13
FORMULA_NAME Angiogenesis-PT-10-CTLFireability-14
FORMULA_NAME Angiogenesis-PT-10-CTLFireability-15
FORMULA_NAME Angiogenesis-PT-10-CTLFireability-2
FORMULA_NAME Angiogenesis-PT-10-CTLFireability-3
FORMULA_NAME Angiogenesis-PT-10-CTLFireability-4
FORMULA_NAME Angiogenesis-PT-10-CTLFireability-5
FORMULA_NAME Angiogenesis-PT-10-CTLFireability-6
FORMULA_NAME Angiogenesis-PT-10-CTLFireability-7
FORMULA_NAME Angiogenesis-PT-10-CTLFireability-8
FORMULA_NAME Angiogenesis-PT-10-CTLFireability-9
=== Now, execution of the tool begins
BK_START 1463512796895
FORMULA Angiogenesis-PT-10-CTLFireability-0 TRUE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN STATIC_VARIABLE_REORDERING
FORMULA Angiogenesis-PT-10-CTLFireability-1 TRUE TECHNIQUES DECISION_DIAGRAMS PARALLEL_PROCESSING USE_NUPN STATIC_VARIABLE_REORDERING
FORMULA Angiogenesis-PT-10-CTLFireability-2 CANNOT_COMPUTE
FORMULA Angiogenesis-PT-10-CTLFireability-3 CANNOT_COMPUTE
FORMULA Angiogenesis-PT-10-CTLFireability-4 CANNOT_COMPUTE
FORMULA Angiogenesis-PT-10-CTLFireability-5 CANNOT_COMPUTE
FORMULA Angiogenesis-PT-10-CTLFireability-6 CANNOT_COMPUTE
FORMULA Angiogenesis-PT-10-CTLFireability-7 CANNOT_COMPUTE
FORMULA Angiogenesis-PT-10-CTLFireability-8 CANNOT_COMPUTE
FORMULA Angiogenesis-PT-10-CTLFireability-9 CANNOT_COMPUTE
FORMULA Angiogenesis-PT-10-CTLFireability-10 CANNOT_COMPUTE
FORMULA Angiogenesis-PT-10-CTLFireability-11 CANNOT_COMPUTE
FORMULA Angiogenesis-PT-10-CTLFireability-12 CANNOT_COMPUTE
FORMULA Angiogenesis-PT-10-CTLFireability-13 CANNOT_COMPUTE
FORMULA Angiogenesis-PT-10-CTLFireability-14 CANNOT_COMPUTE
FORMULA Angiogenesis-PT-10-CTLFireability-15 CANNOT_COMPUTE
BK_STOP 1463516369429
--------------------
content from stderr:
ctl formula name Angiogenesis-PT-10-CTLFireability-0
ctl formula formula --ctl=/tmp/ctl_0_
ctl formula name Angiogenesis-PT-10-CTLFireability-1
ctl formula formula --ctl=/tmp/ctl_1_
ctl formula name Angiogenesis-PT-10-CTLFireability-2
ctl formula formula --ctl=/tmp/ctl_2_
ctl formula name Angiogenesis-PT-10-CTLFireability-3
ctl formula formula --ctl=/tmp/ctl_3_
ctl formula name Angiogenesis-PT-10-CTLFireability-4
ctl formula formula --ctl=/tmp/ctl_4_
ctl formula name Angiogenesis-PT-10-CTLFireability-5
ctl formula formula --ctl=/tmp/ctl_5_
ctl formula name Angiogenesis-PT-10-CTLFireability-6
ctl formula formula --ctl=/tmp/ctl_6_
ctl formula name Angiogenesis-PT-10-CTLFireability-7
ctl formula formula --ctl=/tmp/ctl_7_
ctl formula name Angiogenesis-PT-10-CTLFireability-8
ctl formula formula --ctl=/tmp/ctl_8_
ctl formula name Angiogenesis-PT-10-CTLFireability-9
ctl formula formula --ctl=/tmp/ctl_9_
ctl formula name Angiogenesis-PT-10-CTLFireability-10
ctl formula formula --ctl=/tmp/ctl_10_
ctl formula name Angiogenesis-PT-10-CTLFireability-11
ctl formula formula --ctl=/tmp/ctl_11_
ctl formula name Angiogenesis-PT-10-CTLFireability-12
ctl formula formula --ctl=/tmp/ctl_12_
ctl formula name Angiogenesis-PT-10-CTLFireability-13
ctl formula formula --ctl=/tmp/ctl_13_
ctl formula name Angiogenesis-PT-10-CTLFireability-14
ctl formula formula --ctl=/tmp/ctl_14_
ctl formula name Angiogenesis-PT-10-CTLFireability-15
ctl formula formula --ctl=/tmp/ctl_15_
pnml2lts-sym: Exploration order is chain
pnml2lts-sym: Saturation strategy is sat-like
pnml2lts-sym: Guided search strategy is unguided
pnml2lts-sym: Attractor strategy is default
pnml2lts-sym: opening model.pnml
pnml2lts-sym: Petri net has 39 places, 64 transitions and 185 arcs
pnml2lts-sym: There are 0 safe places
pnml2lts-sym: Petri net model.pnml analyzed
pnml2lts-sym: Loading Petri net took 0.010 real 0.000 user 0.000 sys
pnml2lts-sym: Initializing regrouping layer
pnml2lts-sym: Regroup specification: bs,w2W,ru,hf
pnml2lts-sym: Regroup Boost's Sloan
pnml2lts-sym: Regroup over-approximate must-write to may-write
pnml2lts-sym: Regroup Row sUbsume
pnml2lts-sym: Reqroup Horizontal Flip
pnml2lts-sym: Regrouping: 64->40 groups
pnml2lts-sym: Regrouping took 0.000 real 0.000 user 0.010 sys
pnml2lts-sym: state vector length is 39; there are 40 groups
pnml2lts-sym: Creating a multi-core ListDD domain.
pnml2lts-sym: Using GBgetTransitionsShortR2W as next-state function
pnml2lts-sym: got initial state
pnml2lts-sym: parsing CTL formula
pnml2lts-sym: converting CTL to mu-calculus...
pnml2lts-sym: parsing CTL formula
pnml2lts-sym: converting CTL to mu-calculus...
pnml2lts-sym: parsing CTL formula
pnml2lts-sym: converting CTL to mu-calculus...
pnml2lts-sym: parsing CTL formula
pnml2lts-sym: converting CTL to mu-calculus...
pnml2lts-sym: parsing CTL formula
pnml2lts-sym: converting CTL to mu-calculus...
pnml2lts-sym: parsing CTL formula
pnml2lts-sym: converting CTL to mu-calculus...
pnml2lts-sym: parsing CTL formula
pnml2lts-sym: converting CTL to mu-calculus...
pnml2lts-sym: parsing CTL formula
pnml2lts-sym: converting CTL to mu-calculus...
pnml2lts-sym: parsing CTL formula
pnml2lts-sym: converting CTL to mu-calculus...
pnml2lts-sym: parsing CTL formula
pnml2lts-sym: converting CTL to mu-calculus...
pnml2lts-sym: parsing CTL formula
pnml2lts-sym: converting CTL to mu-calculus...
pnml2lts-sym: parsing CTL formula
pnml2lts-sym: converting CTL to mu-calculus...
pnml2lts-sym: parsing CTL formula
pnml2lts-sym: converting CTL to mu-calculus...
pnml2lts-sym: parsing CTL formula
pnml2lts-sym: converting CTL to mu-calculus...
pnml2lts-sym: parsing CTL formula
pnml2lts-sym: converting CTL to mu-calculus...
pnml2lts-sym: parsing CTL formula
pnml2lts-sym: converting CTL to mu-calculus...
pnml2lts-sym: vrel_add_act not supported; falling back to vrel_add_cpy
pnml2lts-sym: Exploration took 2901 group checks and 2901 next state calls
pnml2lts-sym: reachability took 103.200 real 359.080 user 52.670 sys
pnml2lts-sym: counting visited states...
pnml2lts-sym: counting took 0.180 real 0.730 user 0.000 sys
pnml2lts-sym: state space has 822645885495 states, 148403 nodes
pnml2lts-sym: Formula /tmp/ctl_0_ holds for the initial state
pnml2lts-sym: Formula /tmp/ctl_1_ holds for the initial state
Sequence of Actions to be Executed by the VM
This is useful if one wants to reexecute the tool in the VM from the submitted image disk.
set -x
# this is for BenchKit: configuration of major elements for the test
export BK_INPUT="Angiogenesis-PT-10"
export BK_EXAMINATION="CTLFireability"
export BK_TOOL="ltsmin"
export BK_RESULT_DIR="/users/gast00/fkordon/BK_RESULTS/OUTPUTS"
export BK_TIME_CONFINEMENT="3600"
export BK_MEMORY_CONFINEMENT="16384"
# this is specific to your benchmark or test
export BIN_DIR="$HOME/BenchKit/bin"
# remove the execution directoty if it exists (to avoid increse of .vmdk images)
if [ -d execution ] ; then
rm -rf execution
fi
tar xzf /home/mcc/BenchKit/INPUTS/Angiogenesis-PT-10.tgz
mv Angiogenesis-PT-10 execution
# this is for BenchKit: explicit launching of the test
cd execution
echo "====================================================================="
echo " Generated by BenchKit 2-2979"
echo " Executing tool ltsmin"
echo " Input is Angiogenesis-PT-10, examination is CTLFireability"
echo " Time confinement is $BK_TIME_CONFINEMENT seconds"
echo " Memory confinement is 16384 MBytes"
echo " Number of cores is 4"
echo " Run identifier is r003kn-ebro-146347663700031"
echo "====================================================================="
echo
echo "--------------------"
echo "content from stdout:"
echo
echo "=== Data for post analysis generated by BenchKit (invocation template)"
echo
if [ "CTLFireability" = "UpperBounds" ] ; then
echo "The expected result is a vector of positive values"
echo NUM_VECTOR
elif [ "CTLFireability" != "StateSpace" ] ; then
echo "The expected result is a vector of booleans"
echo BOOL_VECTOR
else
echo "no data necessary for post analysis"
fi
echo
if [ -f "CTLFireability.txt" ] ; then
echo "here is the order used to build the result vector(from text file)"
for x in $(grep Property CTLFireability.txt | cut -d ' ' -f 2 | sort -u) ; do
echo "FORMULA_NAME $x"
done
elif [ -f "CTLFireability.xml" ] ; then # for cunf (txt files deleted;-)
echo echo "here is the order used to build the result vector(from xml file)"
for x in $(grep '
echo "FORMULA_NAME $x"
done
fi
echo
echo "=== Now, execution of the tool begins"
echo
echo -n "BK_START "
date -u +%s%3N
echo
timeout -s 9 $BK_TIME_CONFINEMENT bash -c "/home/mcc/BenchKit/BenchKit_head.sh 2> STDERR ; echo ; echo -n \"BK_STOP \" ; date -u +%s%3N"
if [ $? -eq 137 ] ; then
echo
echo "BK_TIME_CONFINEMENT_REACHED"
fi
echo
echo "--------------------"
echo "content from stderr:"
echo
cat STDERR ;

