About the Execution of Marcie for S_Angiogenesis-PT-10
| Execution Summary | |||||
| Max Memory Used (MB) |
Time wait (ms) | CPU Usage (ms) | I/O Wait (ms) | Computed Result | Execution Status |
| 10288.700 | 1620817.00 | 1620889.00 | 20.00 | TFFFFTTTTTTTTTTT | normal |
Execution Chart
We display below the execution chart for this examination (boot time has been removed).
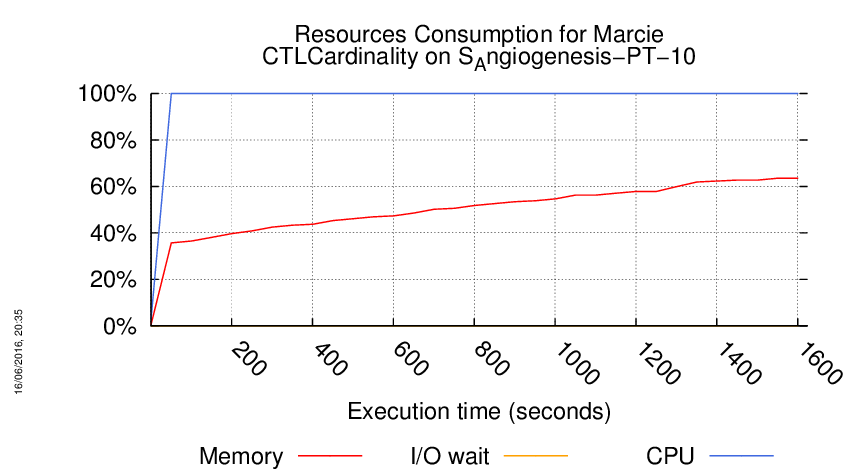
Trace from the execution
Waiting for the VM to be ready (probing ssh)
................
=====================================================================
Generated by BenchKit 2-2979
Executing tool marcie
Input is S_Angiogenesis-PT-10, examination is CTLCardinality
Time confinement is 3600 seconds
Memory confinement is 16384 MBytes
Number of cores is 1
Run identifier is r173kn-ebro-146433144700030
=====================================================================
--------------------
content from stdout:
=== Data for post analysis generated by BenchKit (invocation template)
The expected result is a vector of booleans
BOOL_VECTOR
here is the order used to build the result vector(from text file)
FORMULA_NAME Angiogenesis-PT-10-CTLCardinality-0
FORMULA_NAME Angiogenesis-PT-10-CTLCardinality-1
FORMULA_NAME Angiogenesis-PT-10-CTLCardinality-10
FORMULA_NAME Angiogenesis-PT-10-CTLCardinality-11
FORMULA_NAME Angiogenesis-PT-10-CTLCardinality-12
FORMULA_NAME Angiogenesis-PT-10-CTLCardinality-13
FORMULA_NAME Angiogenesis-PT-10-CTLCardinality-14
FORMULA_NAME Angiogenesis-PT-10-CTLCardinality-15
FORMULA_NAME Angiogenesis-PT-10-CTLCardinality-2
FORMULA_NAME Angiogenesis-PT-10-CTLCardinality-3
FORMULA_NAME Angiogenesis-PT-10-CTLCardinality-4
FORMULA_NAME Angiogenesis-PT-10-CTLCardinality-5
FORMULA_NAME Angiogenesis-PT-10-CTLCardinality-6
FORMULA_NAME Angiogenesis-PT-10-CTLCardinality-7
FORMULA_NAME Angiogenesis-PT-10-CTLCardinality-8
FORMULA_NAME Angiogenesis-PT-10-CTLCardinality-9
=== Now, execution of the tool begins
BK_START 1464784852165
Marcie rev. 8535M (built: crohr on 2016-04-27)
A model checker for Generalized Stochastic Petri nets
authors: Alex Tovchigrechko (IDD package and CTL model checking)
Martin Schwarick (Symbolic numerical analysis and CSL model checking)
Christian Rohr (Simulative and approximative numerical model checking)
marcie@informatik.tu-cottbus.de
called as: marcie --net-file=model.pnml --mcc-file=CTLCardinality.xml --mcc-mode --memory=6 --suppress
parse successfull
net created successfully
Net: Angiogenesis_PT_10
(NrP: 39 NrTr: 64 NrArc: 185)
net check time: 0m 0.002sec
parse formulas
formulas created successfully
place and transition orderings generation:0m 0.000sec
init dd package: 0m 6.064sec
RS generation: 0m16.402sec
-> reachability set: #nodes 128274 (1.3e+05) #states 822,645,885,495 (11)
starting MCC model checker
--------------------------
checking: AX [EF [3<=KdStarGStarP3]]
normalized: ~ [EX [~ [E [true U 3<=KdStarGStarP3]]]]
abstracting: (3<=KdStarGStarP3) states: 4,983,054,645 (9)
.-> the formula is TRUE
FORMULA Angiogenesis-PT-10-CTLCardinality-0 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 1m17.471sec
checking: EF [3<=KdStarGStarPgStar]
normalized: E [true U 3<=KdStarGStarPgStar]
abstracting: (3<=KdStarGStarPgStar) states: 53,746,751,025 (10)
-> the formula is TRUE
FORMULA Angiogenesis-PT-10-CTLCardinality-3 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m30.064sec
checking: AG [AG [~ [3<=KdStarGStarP3]]]
normalized: ~ [E [true U E [true U 3<=KdStarGStarP3]]]
abstracting: (3<=KdStarGStarP3) states: 4,983,054,645 (9)
-> the formula is FALSE
FORMULA Angiogenesis-PT-10-CTLCardinality-12 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 1.190sec
checking: EF [~ [AX [2<=P3k]]]
normalized: E [true U EX [~ [2<=P3k]]]
abstracting: (2<=P3k) states: 821,813,315,242 (11)
.-> the formula is TRUE
FORMULA Angiogenesis-PT-10-CTLCardinality-14 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m59.277sec
checking: ~ [EF [~ [KdStarGStarP3<=KdStarGStar]]]
normalized: ~ [E [true U ~ [KdStarGStarP3<=KdStarGStar]]]
abstracting: (KdStarGStarP3<=KdStarGStar) states: 716,738,019,993 (11)
-> the formula is FALSE
FORMULA Angiogenesis-PT-10-CTLCardinality-10 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m54.840sec
checking: EF [EX [[3<=GStarP3 & 1<=KdStarPgStarP2]]]
normalized: E [true U EX [[3<=GStarP3 & 1<=KdStarPgStarP2]]]
abstracting: (1<=KdStarPgStarP2) states: 209,049,404,819 (11)
abstracting: (3<=GStarP3) states: 20,159,352,225 (10)
.-> the formula is TRUE
FORMULA Angiogenesis-PT-10-CTLCardinality-15 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 1m27.532sec
checking: EF [~ [[[2<=Enz | 2<=KdStarPgStarP2] & ~ [2<=DAGE]]]]
normalized: E [true U ~ [[~ [2<=DAGE] & [2<=Enz | 2<=KdStarPgStarP2]]]]
abstracting: (2<=KdStarPgStarP2) states: 44,154,459,816 (10)
abstracting: (2<=Enz) states: 822,638,391,349 (11)
abstracting: (2<=DAGE) states: 153,496,099,547 (11)
-> the formula is TRUE
FORMULA Angiogenesis-PT-10-CTLCardinality-4 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 7.342sec
checking: AF [EF [[KdStarGStarP3kStar<=KdStarGStarP3kStarP3P2 & 2<=KdStarGStarPgStarP2]]]
normalized: ~ [EG [~ [E [true U [KdStarGStarP3kStar<=KdStarGStarP3kStarP3P2 & 2<=KdStarGStarPgStarP2]]]]]
abstracting: (2<=KdStarGStarPgStarP2) states: 30,441,494,561 (10)
abstracting: (KdStarGStarP3kStar<=KdStarGStarP3kStarP3P2) states: 476,651,324,403 (11)
.
EG iterations: 1
-> the formula is TRUE
FORMULA Angiogenesis-PT-10-CTLCardinality-6 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m57.700sec
checking: E [AX [KdStarGStarP3kStar<=DAG] U AX [2<=KdStarPgStarP2]]
normalized: E [~ [EX [~ [KdStarGStarP3kStar<=DAG]]] U ~ [EX [~ [2<=KdStarPgStarP2]]]]
abstracting: (2<=KdStarPgStarP2) states: 44,154,459,816 (10)
.abstracting: (KdStarGStarP3kStar<=DAG) states: 577,556,125,711 (11)
.-> the formula is TRUE
FORMULA Angiogenesis-PT-10-CTLCardinality-9 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 3m26.490sec
checking: AG [AG [[KdStarGStarPgStarP3P2<=GStarP3kP3 & 3<=Gab1]]]
normalized: ~ [E [true U E [true U ~ [[KdStarGStarPgStarP3P2<=GStarP3kP3 & 3<=Gab1]]]]]
abstracting: (3<=Gab1) states: 222,635,590,715 (11)
abstracting: (KdStarGStarPgStarP3P2<=GStarP3kP3) states: 771,503,483,406 (11)
-> the formula is FALSE
FORMULA Angiogenesis-PT-10-CTLCardinality-11 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m39.828sec
checking: E [[~ [3<=KdStarGStarP3] | [2<=GP3 & 3<=PtP3P2]] U AF [2<=KdStarGStarP3]]
normalized: E [[[2<=GP3 & 3<=PtP3P2] | ~ [3<=KdStarGStarP3]] U ~ [EG [~ [2<=KdStarGStarP3]]]]
abstracting: (2<=KdStarGStarP3) states: 30,725,412,017 (10)
.
EG iterations: 1
abstracting: (3<=KdStarGStarP3) states: 4,983,054,645 (9)
abstracting: (3<=PtP3P2) states: 1,634,615,504 (9)
abstracting: (2<=GP3) states: 80,100,980,848 (10)
-> the formula is TRUE
FORMULA Angiogenesis-PT-10-CTLCardinality-2 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 1m47.909sec
checking: [~ [[3<=KdStarGStarP3k | AF [KdStarG<=KdStarGStarPgStar]]] | ~ [~ [EF [3<=KdStarGStarPgStarP3]]]]
normalized: [E [true U 3<=KdStarGStarPgStarP3] | ~ [[3<=KdStarGStarP3k | ~ [EG [~ [KdStarG<=KdStarGStarPgStar]]]]]]
abstracting: (KdStarG<=KdStarGStarPgStar) states: 564,983,470,496 (11)
.
EG iterations: 1
abstracting: (3<=KdStarGStarP3k) states: 54,659,434,847 (10)
abstracting: (3<=KdStarGStarPgStarP3) states: 4,926,076,264 (9)
-> the formula is TRUE
FORMULA Angiogenesis-PT-10-CTLCardinality-5 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 1m33.498sec
checking: AG [[AG [Gab1<=KdStarPgStar] | EF [KdStarGStarPg<=GStarPgP3]]]
normalized: ~ [E [true U ~ [[E [true U KdStarGStarPg<=GStarPgP3] | ~ [E [true U ~ [Gab1<=KdStarPgStar]]]]]]]
abstracting: (Gab1<=KdStarPgStar) states: 421,259,131,327 (11)
abstracting: (KdStarGStarPg<=GStarPgP3) states: 538,649,769,753 (11)
-> the formula is TRUE
FORMULA Angiogenesis-PT-10-CTLCardinality-7 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 2m55.577sec
checking: AX [EF [[Gab1<=KdStarGStarPgStar & KdStarGStarP3<=KdStarGStarP3kP3]]]
normalized: ~ [EX [~ [E [true U [Gab1<=KdStarGStarPgStar & KdStarGStarP3<=KdStarGStarP3kP3]]]]]
abstracting: (KdStarGStarP3<=KdStarGStarP3kP3) states: 681,258,533,230 (11)
abstracting: (Gab1<=KdStarGStarPgStar) states: 356,466,615,377 (11)
.-> the formula is TRUE
FORMULA Angiogenesis-PT-10-CTLCardinality-8 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 1m47.007sec
checking: ~ [[EF [3<=KdStarGStarPg] & AG [[KdStarPgStar<=Gab1 & KdStarGStarP3kP3<=PtP3]]]]
normalized: ~ [[~ [E [true U ~ [[KdStarPgStar<=Gab1 & KdStarGStarP3kP3<=PtP3]]]] & E [true U 3<=KdStarGStarPg]]]
abstracting: (3<=KdStarGStarPg) states: 53,746,751,025 (10)
abstracting: (KdStarGStarP3kP3<=PtP3) states: 718,094,056,771 (11)
abstracting: (KdStarPgStar<=Gab1) states: 633,233,668,264 (11)
-> the formula is TRUE
FORMULA Angiogenesis-PT-10-CTLCardinality-13 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 5m50.080sec
checking: ~ [[EF [[2<=KdStarGStarPg & 2<=KdStarGStarP3kStar]] & [[~ [2<=KdStarPgStar] | [2<=PtP3 | 3<=Pip3]] | [~ [KdStarPgStarP2<=GStarP3] & [1<=KdStarGStarPgStarP3 & 2<=Enz]]]]]
normalized: ~ [[[[~ [KdStarPgStarP2<=GStarP3] & [1<=KdStarGStarPgStarP3 & 2<=Enz]] | [[2<=PtP3 | 3<=Pip3] | ~ [2<=KdStarPgStar]]] & E [true U [2<=KdStarGStarPg & 2<=KdStarGStarP3kStar]]]]
abstracting: (2<=KdStarGStarP3kStar) states: 148,603,578,929 (11)
abstracting: (2<=KdStarGStarPg) states: 146,837,300,519 (11)
abstracting: (2<=KdStarPgStar) states: 237,492,609,082 (11)
abstracting: (3<=Pip3) states: 58,569,550,836 (10)
abstracting: (2<=PtP3) states: 153,496,000,877 (11)
abstracting: (2<=Enz) states: 822,638,391,349 (11)
abstracting: (1<=KdStarGStarPgStarP3) states: 166,727,081,996 (11)
abstracting: (KdStarPgStarP2<=GStarP3) states: 673,543,465,088 (11)
-> the formula is FALSE
FORMULA Angiogenesis-PT-10-CTLCardinality-1 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 2m 0.687sec
Total processing time: 27m 0.397sec
BK_STOP 1464786472982
--------------------
content from stderr:
check for maximal unmarked siphon
ok
check if there are places and transitions
ok
check if there are transitions without pre-places
ok
check if at least one transition is enabled in m0
ok
check if there are transitions that can never fire
ok
initing FirstDep: 0m 0.000sec
3968 3382 5295 11703 17108 31307 124827 133681 131288 129623 128662
iterations count:11953 (186), effective:901 (14)
initing FirstDep: 0m 0.000sec
147248 163067 188467 169570 147006
iterations count:5166 (80), effective:388 (6)
149474 152814 149793
iterations count:3061 (47), effective:228 (3)
147248 163067 188467 169570 147006
iterations count:5166 (80), effective:388 (6)
iterations count:64 (1), effective:0 (0)
41861 71151 77977 77181 104938 121799 153202 129150 128255 139430 143969
iterations count:11995 (187), effective:973 (15)
146885
iterations count:1512 (23), effective:129 (2)
107040 137273 141352 143086 130471
iterations count:5043 (78), effective:390 (6)
121347 133151
iterations count:2357 (36), effective:171 (2)
66756 79629 107571 166306 169242
iterations count:5557 (86), effective:461 (7)
52550 61716 64492 62404 153423 250089 372509 370017 371174 289641 252709 232706 211255
iterations count:13052 (203), effective:1048 (16)
131926 130817 138564
iterations count:3592 (56), effective:296 (4)
iterations count:64 (1), effective:0 (0)
145667 168995 146995
iterations count:3071 (47), effective:225 (3)
123515 181093 187492
iterations count:3869 (60), effective:289 (4)
205566 174312 156131 143992
iterations count:4690 (73), effective:354 (5)
iterations count:707 (11), effective:61 (0)
141606 131664 135962 136743 136326 136582 140361 140691 140762
iterations count:9540 (149), effective:765 (11)
136030 132808 145154 145401
iterations count:4979 (77), effective:380 (5)
245922 129795
iterations count:2433 (38), effective:208 (3)
87996 119555 135581 145827 146644 147290
iterations count:6875 (107), effective:524 (8)
Sequence of Actions to be Executed by the VM
This is useful if one wants to reexecute the tool in the VM from the submitted image disk.
set -x
# this is for BenchKit: configuration of major elements for the test
export BK_INPUT="S_Angiogenesis-PT-10"
export BK_EXAMINATION="CTLCardinality"
export BK_TOOL="marcie"
export BK_RESULT_DIR="/users/gast00/fkordon/BK_RESULTS/OUTPUTS"
export BK_TIME_CONFINEMENT="3600"
export BK_MEMORY_CONFINEMENT="16384"
# this is specific to your benchmark or test
export BIN_DIR="$HOME/BenchKit/bin"
# remove the execution directoty if it exists (to avoid increse of .vmdk images)
if [ -d execution ] ; then
rm -rf execution
fi
tar xzf /home/mcc/BenchKit/INPUTS/S_Angiogenesis-PT-10.tgz
mv S_Angiogenesis-PT-10 execution
# this is for BenchKit: explicit launching of the test
cd execution
echo "====================================================================="
echo " Generated by BenchKit 2-2979"
echo " Executing tool marcie"
echo " Input is S_Angiogenesis-PT-10, examination is CTLCardinality"
echo " Time confinement is $BK_TIME_CONFINEMENT seconds"
echo " Memory confinement is 16384 MBytes"
echo " Number of cores is 1"
echo " Run identifier is r173kn-ebro-146433144700030"
echo "====================================================================="
echo
echo "--------------------"
echo "content from stdout:"
echo
echo "=== Data for post analysis generated by BenchKit (invocation template)"
echo
if [ "CTLCardinality" = "UpperBounds" ] ; then
echo "The expected result is a vector of positive values"
echo NUM_VECTOR
elif [ "CTLCardinality" != "StateSpace" ] ; then
echo "The expected result is a vector of booleans"
echo BOOL_VECTOR
else
echo "no data necessary for post analysis"
fi
echo
if [ -f "CTLCardinality.txt" ] ; then
echo "here is the order used to build the result vector(from text file)"
for x in $(grep Property CTLCardinality.txt | cut -d ' ' -f 2 | sort -u) ; do
echo "FORMULA_NAME $x"
done
elif [ -f "CTLCardinality.xml" ] ; then # for cunf (txt files deleted;-)
echo echo "here is the order used to build the result vector(from xml file)"
for x in $(grep '
echo "FORMULA_NAME $x"
done
fi
echo
echo "=== Now, execution of the tool begins"
echo
echo -n "BK_START "
date -u +%s%3N
echo
timeout -s 9 $BK_TIME_CONFINEMENT bash -c "/home/mcc/BenchKit/BenchKit_head.sh 2> STDERR ; echo ; echo -n \"BK_STOP \" ; date -u +%s%3N"
if [ $? -eq 137 ] ; then
echo
echo "BK_TIME_CONFINEMENT_REACHED"
fi
echo
echo "--------------------"
echo "content from stderr:"
echo
cat STDERR ;

