About the Execution of Marcie for Angiogenesis-PT-05
| Execution Summary | |||||
| Max Memory Used (MB) |
Time wait (ms) | CPU Usage (ms) | I/O Wait (ms) | Computed Result | Execution Status |
| 5610.700 | 38504.00 | 38601.00 | 50.30 | FFTTFTTTFTFTFTFT | normal |
Execution Chart
We display below the execution chart for this examination (boot time has been removed).
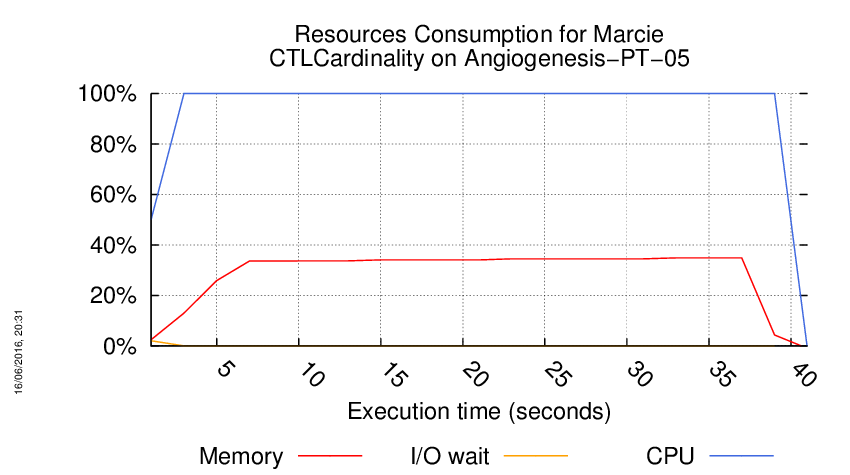
Trace from the execution
Waiting for the VM to be ready (probing ssh)
................
=====================================================================
Generated by BenchKit 2-2979
Executing tool marcie
Input is Angiogenesis-PT-05, examination is CTLCardinality
Time confinement is 3600 seconds
Memory confinement is 16384 MBytes
Number of cores is 1
Run identifier is r005kn-ebro-146347672600021
=====================================================================
--------------------
content from stdout:
=== Data for post analysis generated by BenchKit (invocation template)
The expected result is a vector of booleans
BOOL_VECTOR
here is the order used to build the result vector(from text file)
FORMULA_NAME Angiogenesis-PT-05-CTLCardinality-0
FORMULA_NAME Angiogenesis-PT-05-CTLCardinality-1
FORMULA_NAME Angiogenesis-PT-05-CTLCardinality-10
FORMULA_NAME Angiogenesis-PT-05-CTLCardinality-11
FORMULA_NAME Angiogenesis-PT-05-CTLCardinality-12
FORMULA_NAME Angiogenesis-PT-05-CTLCardinality-13
FORMULA_NAME Angiogenesis-PT-05-CTLCardinality-14
FORMULA_NAME Angiogenesis-PT-05-CTLCardinality-15
FORMULA_NAME Angiogenesis-PT-05-CTLCardinality-2
FORMULA_NAME Angiogenesis-PT-05-CTLCardinality-3
FORMULA_NAME Angiogenesis-PT-05-CTLCardinality-4
FORMULA_NAME Angiogenesis-PT-05-CTLCardinality-5
FORMULA_NAME Angiogenesis-PT-05-CTLCardinality-6
FORMULA_NAME Angiogenesis-PT-05-CTLCardinality-7
FORMULA_NAME Angiogenesis-PT-05-CTLCardinality-8
FORMULA_NAME Angiogenesis-PT-05-CTLCardinality-9
=== Now, execution of the tool begins
BK_START 1463590754317
Marcie rev. 8535M (built: crohr on 2016-04-27)
A model checker for Generalized Stochastic Petri nets
authors: Alex Tovchigrechko (IDD package and CTL model checking)
Martin Schwarick (Symbolic numerical analysis and CSL model checking)
Christian Rohr (Simulative and approximative numerical model checking)
marcie@informatik.tu-cottbus.de
called as: marcie --net-file=model.pnml --mcc-file=CTLCardinality.xml --mcc-mode --memory=6 --suppress
parse successfull
net created successfully
Net: Angiogenesis_PT_05
(NrP: 39 NrTr: 64 NrArc: 185)
net check time: 0m 0.000sec
parse formulas
formulas created successfully
place and transition orderings generation:0m 0.000sec
init dd package: 0m 6.127sec
RS generation: 0m 0.858sec
-> reachability set: #nodes 10921 (1.1e+04) #states 42,734,935 (7)
starting MCC model checker
--------------------------
checking: ~ [EF [3<=KdStarGStarP3kStar]]
normalized: ~ [E [true U 3<=KdStarGStarP3kStar]]
abstracting: (3<=KdStarGStarP3kStar) states: 512,541 (5)
-> the formula is FALSE
FORMULA Angiogenesis-PT-05-CTLCardinality-0 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 1.582sec
checking: A [EX [3<=PtP2] U EF [1<=KdStarGStarPgP3]]
normalized: [~ [EG [~ [E [true U 1<=KdStarGStarPgP3]]]] & ~ [E [~ [E [true U 1<=KdStarGStarPgP3]] U [~ [E [true U 1<=KdStarGStarPgP3]] & ~ [EX [3<=PtP2]]]]]]
abstracting: (3<=PtP2) states: 719,420 (5)
.abstracting: (1<=KdStarGStarPgP3) states: 4,166,196 (6)
abstracting: (1<=KdStarGStarPgP3) states: 4,166,196 (6)
abstracting: (1<=KdStarGStarPgP3) states: 4,166,196 (6)
.
EG iterations: 1
-> the formula is TRUE
FORMULA Angiogenesis-PT-05-CTLCardinality-3 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 1.231sec
checking: AG [EG [[1<=KdStarGStarPg | 3<=Akt]]]
normalized: ~ [E [true U ~ [EG [[1<=KdStarGStarPg | 3<=Akt]]]]]
abstracting: (3<=Akt) states: 19,822,700 (7)
abstracting: (1<=KdStarGStarPg) states: 12,421,596 (7)
.
EG iterations: 1
-> the formula is FALSE
FORMULA Angiogenesis-PT-05-CTLCardinality-4 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 2.452sec
checking: AG [EX [[3<=KdStarGStarPgStarP3 | 1<=P3k]]]
normalized: ~ [E [true U ~ [EX [[3<=KdStarGStarPgStarP3 | 1<=P3k]]]]]
abstracting: (1<=P3k) states: 42,575,177 (7)
abstracting: (3<=KdStarGStarPgStarP3) states: 15,826 (4)
.-> the formula is FALSE
FORMULA Angiogenesis-PT-05-CTLCardinality-12 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.913sec
checking: AG [Gab1<=Gab1]
normalized: ~ [E [true U ~ [Gab1<=Gab1]]]
abstracting: (Gab1<=Gab1) states: 42,734,935 (7)
-> the formula is TRUE
FORMULA Angiogenesis-PT-05-CTLCardinality-14 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.020sec
checking: [EF [~ [3<=KdStarPg]] & ~ [EF [[2<=AktStar & 2<=KdStarPgStarP2]]]]
normalized: [~ [E [true U [2<=AktStar & 2<=KdStarPgStarP2]]] & E [true U ~ [3<=KdStarPg]]]
abstracting: (3<=KdStarPg) states: 1,170,290 (6)
abstracting: (2<=KdStarPgStarP2) states: 433,155 (5)
abstracting: (2<=AktStar) states: 27,444,286 (7)
-> the formula is FALSE
FORMULA Angiogenesis-PT-05-CTLCardinality-1 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 1.691sec
checking: AG [E [PtP2<=PtP2 U 1<=KdStarGStarP3kStarP2]]
normalized: ~ [E [true U ~ [E [PtP2<=PtP2 U 1<=KdStarGStarP3kStarP2]]]]
abstracting: (1<=KdStarGStarP3kStarP2) states: 4,240,836 (6)
abstracting: (PtP2<=PtP2) states: 42,734,935 (7)
-> the formula is FALSE
FORMULA Angiogenesis-PT-05-CTLCardinality-2 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 1.037sec
checking: ~ [[~ [EF [3<=KdStarGStarPgStarP2]] & E [PtP3P2<=AktStar U 2<=KdStarGStarP3kStarP2]]]
normalized: ~ [[E [PtP3P2<=AktStar U 2<=KdStarGStarP3kStarP2] & ~ [E [true U 3<=KdStarGStarPgStarP2]]]]
abstracting: (3<=KdStarGStarPgStarP2) states: 15,826 (4)
abstracting: (2<=KdStarGStarP3kStarP2) states: 314,760 (5)
abstracting: (PtP3P2<=AktStar) states: 42,105,035 (7)
-> the formula is TRUE
FORMULA Angiogenesis-PT-05-CTLCardinality-5 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 2.086sec
checking: AG [EF [[P3k<=DAG & KdStarPgStar<=KdStarGStar]]]
normalized: ~ [E [true U ~ [E [true U [P3k<=DAG & KdStarPgStar<=KdStarGStar]]]]]
abstracting: (KdStarPgStar<=KdStarGStar) states: 28,956,633 (7)
abstracting: (P3k<=DAG) states: 1,114,608 (6)
-> the formula is FALSE
FORMULA Angiogenesis-PT-05-CTLCardinality-6 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 2.500sec
checking: E [~ [[3<=KdStarGStarPgStarP2 & GP3<=P3k]] U ~ [~ [3<=KdStarGStarP3kStarP2]]]
normalized: E [~ [[3<=KdStarGStarPgStarP2 & GP3<=P3k]] U 3<=KdStarGStarP3kStarP2]
abstracting: (3<=KdStarGStarP3kStarP2) states: 16,257 (4)
abstracting: (GP3<=P3k) states: 42,734,935 (7)
abstracting: (3<=KdStarGStarPgStarP2) states: 15,826 (4)
-> the formula is TRUE
FORMULA Angiogenesis-PT-05-CTLCardinality-11 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.561sec
checking: [E [KdStarPgStar<=KdStarGStarPgStar U Enz<=KdStarGStarPgStarP3] & 1<=Pten]
normalized: [1<=Pten & E [KdStarPgStar<=KdStarGStarPgStar U Enz<=KdStarGStarPgStarP3]]
abstracting: (Enz<=KdStarGStarPgStarP3) states: 11,732 (4)
abstracting: (KdStarPgStar<=KdStarGStarPgStar) states: 28,849,941 (7)
abstracting: (1<=Pten) states: 42,690,563 (7)
-> the formula is TRUE
FORMULA Angiogenesis-PT-05-CTLCardinality-9 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 0.278sec
checking: EF [[AX [KdStarGStarP3kStarP3<=AktStar] & [2<=KdStarGStarP3 & KdStar<=KdStarGStarP3kStarP3]]]
normalized: E [true U [~ [EX [~ [KdStarGStarP3kStarP3<=AktStar]]] & [2<=KdStarGStarP3 & KdStar<=KdStarGStarP3kStarP3]]]
abstracting: (KdStar<=KdStarGStarP3kStarP3) states: 26,087,084 (7)
abstracting: (2<=KdStarGStarP3) states: 314,760 (5)
abstracting: (KdStarGStarP3kStarP3<=AktStar) states: 41,927,790 (7)
.-> the formula is TRUE
FORMULA Angiogenesis-PT-05-CTLCardinality-15 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 1.533sec
checking: [E [~ [2<=KdStarPg] U KdStarGStarP3kStarP2<=KdStarGStarP3kP3] & E [~ [Pip2<=AktP3] U 3<=AktStar]]
normalized: [E [~ [Pip2<=AktP3] U 3<=AktStar] & E [~ [2<=KdStarPg] U KdStarGStarP3kStarP2<=KdStarGStarP3kP3]]
abstracting: (KdStarGStarP3kStarP2<=KdStarGStarP3kP3) states: 38,793,091 (7)
abstracting: (2<=KdStarPg) states: 5,349,501 (6)
abstracting: (3<=AktStar) states: 19,817,822 (7)
abstracting: (Pip2<=AktP3) states: 31,613,390 (7)
-> the formula is TRUE
FORMULA Angiogenesis-PT-05-CTLCardinality-7 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 2.682sec
checking: [2<=KdStarGStarP3kStarP3P2 | [[KdStarGStarP3k<=KdStarGStarP3kP3 & EF [Akt<=GStarP3]] | AG [~ [1<=Pg]]]]
normalized: [2<=KdStarGStarP3kStarP3P2 | [~ [E [true U 1<=Pg]] | [KdStarGStarP3k<=KdStarGStarP3kP3 & E [true U Akt<=GStarP3]]]]
abstracting: (Akt<=GStarP3) states: 9,397,523 (6)
abstracting: (KdStarGStarP3k<=KdStarGStarP3kP3) states: 30,915,027 (7)
abstracting: (1<=Pg) states: 39,776,484 (7)
abstracting: (2<=KdStarGStarP3kStarP3P2) states: 11,811 (4)
-> the formula is TRUE
FORMULA Angiogenesis-PT-05-CTLCardinality-13 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 3.588sec
checking: ~ [EF [[[KdStarGStarPgStarP3P2<=KdStarGStarP3kStar | DAGE<=KdStarGStarP3kStar] & [Gab1<=KdStarGStar & Pip2<=GStarP3kP3]]]]
normalized: ~ [E [true U [[Gab1<=KdStarGStar & Pip2<=GStarP3kP3] & [KdStarGStarPgStarP3P2<=KdStarGStarP3kStar | DAGE<=KdStarGStarP3kStar]]]]
abstracting: (DAGE<=KdStarGStarP3kStar) states: 32,542,208 (7)
abstracting: (KdStarGStarPgStarP3P2<=KdStarGStarP3kStar) states: 41,884,847 (7)
abstracting: (Pip2<=GStarP3kP3) states: 31,142,296 (7)
abstracting: (Gab1<=KdStarGStar) states: 21,950,114 (7)
-> the formula is FALSE
FORMULA Angiogenesis-PT-05-CTLCardinality-8 FALSE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 2.453sec
checking: E [[3<=GStarP3 | AktStar<=GStarPgP3] U [[KdStarGStarPgStarP3P2<=P3k | PtP3<=KdStarGStarP3kStar] & [KdStarGStarPgP3<=Pg & 2<=KdStarGStarP3]]]
normalized: E [[3<=GStarP3 | AktStar<=GStarPgP3] U [[KdStarGStarPgStarP3P2<=P3k | PtP3<=KdStarGStarP3kStar] & [KdStarGStarPgP3<=Pg & 2<=KdStarGStarP3]]]
abstracting: (2<=KdStarGStarP3) states: 314,760 (5)
abstracting: (KdStarGStarPgP3<=Pg) states: 42,197,431 (7)
abstracting: (PtP3<=KdStarGStarP3kStar) states: 32,545,112 (7)
abstracting: (KdStarGStarPgStarP3P2<=P3k) states: 42,734,935 (7)
abstracting: (AktStar<=GStarPgP3) states: 9,217,782 (6)
abstracting: (3<=GStarP3) states: 115,195 (5)
-> the formula is TRUE
FORMULA Angiogenesis-PT-05-CTLCardinality-10 TRUE TECHNIQUES SEQUENTIAL_PROCESSING DECISION_DIAGRAMS UNFOLDING_TO_PT
MC time: 0m 3.333sec
Total processing time: 0m38.444sec
BK_STOP 1463590792821
--------------------
content from stderr:
check for maximal unmarked siphon
ok
check if there are places and transitions
ok
check if there are transitions without pre-places
ok
check if at least one transition is enabled in m0
ok
check if there are transitions that can never fire
ok
initing FirstDep: 0m 0.000sec
1096 1888 3952 10799 10915
iterations count:5829 (91), effective:437 (6)
initing FirstDep: 0m 0.000sec
6563 10364 12149 12522
iterations count:4753 (74), effective:351 (5)
12759
iterations count:1061 (16), effective:71 (1)
12759
iterations count:1061 (16), effective:71 (1)
iterations count:64 (1), effective:0 (0)
12759
iterations count:1061 (16), effective:71 (1)
12914
iterations count:1340 (20), effective:95 (1)
2499 3090 4870 7850 10124 11372 11135 12480
iterations count:8609 (134), effective:670 (10)
iterations count:82 (1), effective:3 (0)
6108 14542 16730 15923
iterations count:4873 (76), effective:375 (5)
12727
iterations count:1167 (18), effective:78 (1)
4631 7801 8059 11246
iterations count:4785 (74), effective:390 (6)
3479 10699 14021 13626
iterations count:4678 (73), effective:361 (5)
11082 15389 14563
iterations count:3204 (50), effective:233 (3)
7477 8284 10683 13214 14011 14235 13689 13465
iterations count:8664 (135), effective:690 (10)
4343 6286 8094 11267
iterations count:4871 (76), effective:394 (6)
6579 9662 12455 13828
iterations count:4786 (74), effective:364 (5)
3705 5019 5540 9009 10411
iterations count:5523 (86), effective:430 (6)
7852 12938 12841
iterations count:3528 (55), effective:272 (4)
iterations count:512 (8), effective:44 (0)
16703
iterations count:1910 (29), effective:139 (2)
12061 13113
iterations count:2051 (32), effective:158 (2)
iterations count:561 (8), effective:31 (0)
11987 11813 11835
iterations count:3360 (52), effective:271 (4)
14683 19211 17861
iterations count:3792 (59), effective:284 (4)
Sequence of Actions to be Executed by the VM
This is useful if one wants to reexecute the tool in the VM from the submitted image disk.
set -x
# this is for BenchKit: configuration of major elements for the test
export BK_INPUT="Angiogenesis-PT-05"
export BK_EXAMINATION="CTLCardinality"
export BK_TOOL="marcie"
export BK_RESULT_DIR="/users/gast00/fkordon/BK_RESULTS/OUTPUTS"
export BK_TIME_CONFINEMENT="3600"
export BK_MEMORY_CONFINEMENT="16384"
# this is specific to your benchmark or test
export BIN_DIR="$HOME/BenchKit/bin"
# remove the execution directoty if it exists (to avoid increse of .vmdk images)
if [ -d execution ] ; then
rm -rf execution
fi
tar xzf /home/mcc/BenchKit/INPUTS/Angiogenesis-PT-05.tgz
mv Angiogenesis-PT-05 execution
# this is for BenchKit: explicit launching of the test
cd execution
echo "====================================================================="
echo " Generated by BenchKit 2-2979"
echo " Executing tool marcie"
echo " Input is Angiogenesis-PT-05, examination is CTLCardinality"
echo " Time confinement is $BK_TIME_CONFINEMENT seconds"
echo " Memory confinement is 16384 MBytes"
echo " Number of cores is 1"
echo " Run identifier is r005kn-ebro-146347672600021"
echo "====================================================================="
echo
echo "--------------------"
echo "content from stdout:"
echo
echo "=== Data for post analysis generated by BenchKit (invocation template)"
echo
if [ "CTLCardinality" = "UpperBounds" ] ; then
echo "The expected result is a vector of positive values"
echo NUM_VECTOR
elif [ "CTLCardinality" != "StateSpace" ] ; then
echo "The expected result is a vector of booleans"
echo BOOL_VECTOR
else
echo "no data necessary for post analysis"
fi
echo
if [ -f "CTLCardinality.txt" ] ; then
echo "here is the order used to build the result vector(from text file)"
for x in $(grep Property CTLCardinality.txt | cut -d ' ' -f 2 | sort -u) ; do
echo "FORMULA_NAME $x"
done
elif [ -f "CTLCardinality.xml" ] ; then # for cunf (txt files deleted;-)
echo echo "here is the order used to build the result vector(from xml file)"
for x in $(grep '
echo "FORMULA_NAME $x"
done
fi
echo
echo "=== Now, execution of the tool begins"
echo
echo -n "BK_START "
date -u +%s%3N
echo
timeout -s 9 $BK_TIME_CONFINEMENT bash -c "/home/mcc/BenchKit/BenchKit_head.sh 2> STDERR ; echo ; echo -n \"BK_STOP \" ; date -u +%s%3N"
if [ $? -eq 137 ] ; then
echo
echo "BK_TIME_CONFINEMENT_REACHED"
fi
echo
echo "--------------------"
echo "content from stderr:"
echo
cat STDERR ;

